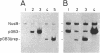
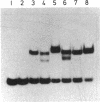
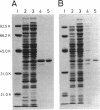
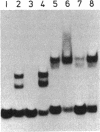
Abstract
The RepA replication protein of plasmid pSC101 binds as a monomer to three repeated sequences (RS1, RS2, and RS3) in the replication origin of the plasmid to initiate duplication and binds as a dimer to two inversely repeated sequences (IR1 and IR2) in its promoter region (D. Manen, L. C. Upegui-Gonzalez, and L. Caro, Proc. Natl. Acad. Sci. USA 89:8923-8927, 1992). The binding to IR2 autoregulates repA transcription (P. Linder, G. Churchward, G. X. Xia, Y. Y. Yu, and L. Caro, J. Mol. Biol. 181:383-393, 1985). A mutation in the protein RepA(cop) that affects a single amino acid increases the plasmid copy number fourfold. In vivo experiments show that, when provided in trans under a foreign promoter, the RepA(cop) protein increases the replication of a plasmid containing the origin of replication without repA, whereas it decreases the repression of its own promoter. In vitro experiments show that the purified RepA(cop) protein binds more efficiently to the repeated sequences within the origin than does RepA and that its binding to these sequences is more specific than that of RepA. Binding to an inversely repeated sequence within the repA promoter gives opposite results: the wild-type protein binds efficiently to that sequence, whereas the mutated protein binds less efficiently and less specifically. Footprint experiments confirmed these results and, in addition, showed a difference in the pattern of protection of the inversely repeated sequences by the mutant protein. Equilibrium binding experiments showed that the formation of protein-probe complexes at increasing concentrations of protein had a sigmoidal shape for binding to RS sequences and a hyperbolic shape for binding to IR sequences. The results, together with earlier work (G.-X. Xia, D. Manen, T. Goebel, P. Linder, G. Churchward, and L. Caro, Mol. Microbiol. 5:631-640, 1991), confirm that the binding of RepA to RS sequences plays a crucial role in the regulation of plasmid replication and that its binding to IR sequences plays a role in the autoregulation of RepA expression. They also demonstrate that the two separate functions of the protein are effected by two different forms of binding to the target sites.
Full text
PDF




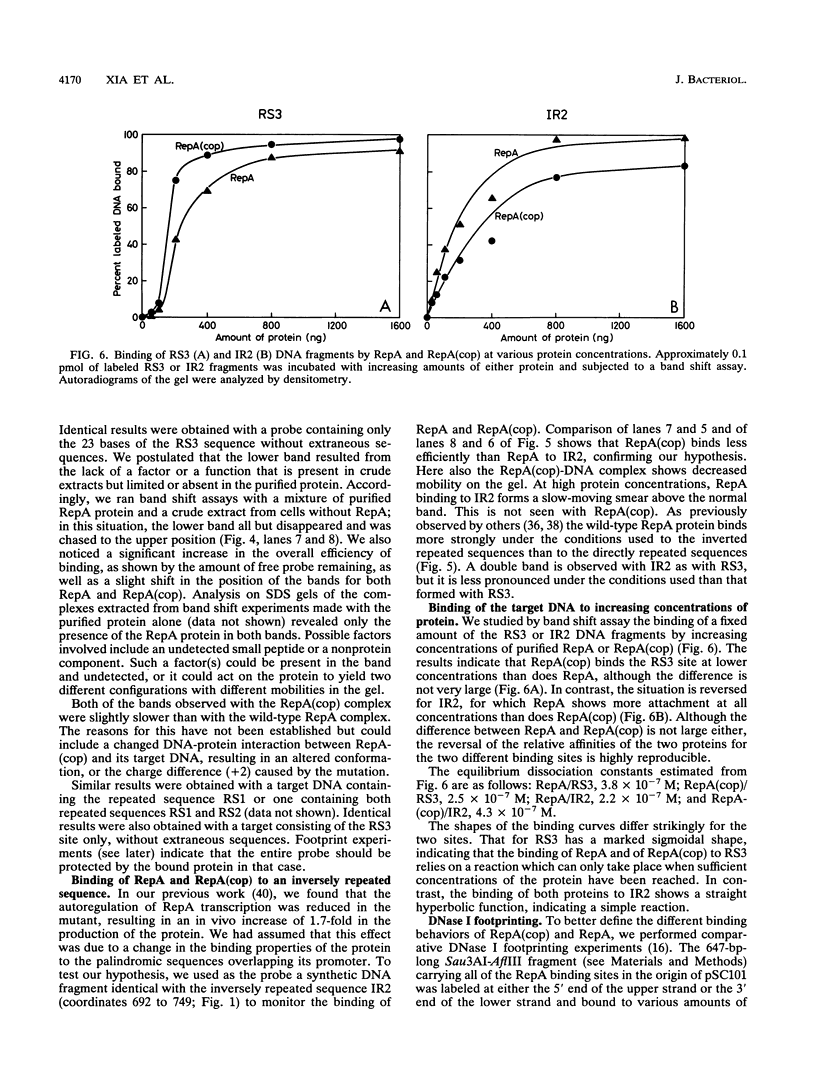



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arini A., Tuscan M., Churchward G. Replication origin mutations affecting binding of pSC101 plasmid-encoded Rep initiator protein. J Bacteriol. 1992 Jan;174(2):456–463. doi: 10.1128/jb.174.2.456-463.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong K. A., Acosta R., Ledner E., Machida Y., Pancotto M., McCormick M., Ohtsubo H., Ohtsubo E. A 37 X 10(3) molecular weight plasmid-encoded protein is required for replication and copy number control in the plasmid pSC101 and its temperature-sensitive derivative pHS1. J Mol Biol. 1984 May 25;175(3):331–348. doi: 10.1016/0022-2836(84)90352-8. [DOI] [PubMed] [Google Scholar]
- Bolivar F., Backman K. Plasmids of Escherichia coli as cloning vectors. Methods Enzymol. 1979;68:245–267. doi: 10.1016/0076-6879(79)68018-7. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Bujard H., Gentz R., Lanzer M., Stueber D., Mueller M., Ibrahimi I., Haeuptle M. T., Dobberstein B. A T5 promoter-based transcription-translation system for the analysis of proteins in vitro and in vivo. Methods Enzymol. 1987;155:416–433. doi: 10.1016/0076-6879(87)55028-5. [DOI] [PubMed] [Google Scholar]
- Bétermier M., Lefrère V., Koch C., Alazard R., Chandler M. The Escherichia coli protein, Fis: specific binding to the ends of phage Mu DNA and modulation of phage growth. Mol Microbiol. 1989 Apr;3(4):459–468. doi: 10.1111/j.1365-2958.1989.tb00192.x. [DOI] [PubMed] [Google Scholar]
- Chang A. C., Cohen S. N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978 Jun;134(3):1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchward G., Belin D., Nagamine Y. A pSC101-derived plasmid which shows no sequence homology to other commonly used cloning vectors. Gene. 1984 Nov;31(1-3):165–171. doi: 10.1016/0378-1119(84)90207-5. [DOI] [PubMed] [Google Scholar]
- Churchward G., Linder P., Caro L. The nucleotide sequence of replication and maintenance functions encoded by plasmid pSC101. Nucleic Acids Res. 1983 Aug 25;11(16):5645–5659. doi: 10.1093/nar/11.16.5645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S. N., Chang A. C. Revised interpretation of the origin of the pSC101 plasmid. J Bacteriol. 1977 Nov;132(2):734–737. doi: 10.1128/jb.132.2.734-737.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durland R. H., Toukdarian A., Fang F., Helinski D. R. Mutations in the trfA replication gene of the broad-host-range plasmid RK2 result in elevated plasmid copy numbers. J Bacteriol. 1990 Jul;172(7):3859–3867. doi: 10.1128/jb.172.7.3859-3867.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fellay R., Frey J., Krisch H. Interposon mutagenesis of soil and water bacteria: a family of DNA fragments designed for in vitro insertional mutagenesis of gram-negative bacteria. Gene. 1987;52(2-3):147–154. doi: 10.1016/0378-1119(87)90041-2. [DOI] [PubMed] [Google Scholar]
- Frey J., Chandler M., Caro L. The effects of an Escherichia coli dnaAts mutation on the replication of the plasmids colE1 pSC101, R100.1 and RTF-TC. Mol Gen Genet. 1979 Jul 13;174(2):117–126. doi: 10.1007/BF00268349. [DOI] [PubMed] [Google Scholar]
- Fried M. G., Crothers D. M. Kinetics and mechanism in the reaction of gene regulatory proteins with DNA. J Mol Biol. 1984 Jan 25;172(3):263–282. doi: 10.1016/s0022-2836(84)80026-1. [DOI] [PubMed] [Google Scholar]
- Galas D. J., Schmitz A. DNAse footprinting: a simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res. 1978 Sep;5(9):3157–3170. doi: 10.1093/nar/5.9.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamas P., Burger A. C., Churchward G., Caro L., Galas D., Chandler M. Replication of pSC101: effects of mutations in the E. coli DNA binding protein IHF. Mol Gen Genet. 1986 Jul;204(1):85–89. doi: 10.1007/BF00330192. [DOI] [PubMed] [Google Scholar]
- Goebel T., Manen D., Alff-Steinberger C., Xia G. X., Caro L. Analysis of a copy number mutant of plasmid pSC101: co-maintenance of wild type and mutant plasmids. Res Microbiol. 1991 Feb-Apr;142(2-3):141–149. doi: 10.1016/0923-2508(91)90022-3. [DOI] [PubMed] [Google Scholar]
- Hasunuma K., Sekiguchi M. Replication of plasmid pSC101 in Escherichia coli K12: requirement for dnaA function. Mol Gen Genet. 1977 Sep 9;154(3):225–230. doi: 10.1007/BF00571277. [DOI] [PubMed] [Google Scholar]
- Kanehisa M. I. Los Alamos sequence analysis package for nucleic acids and proteins. Nucleic Acids Res. 1982 Jan 11;10(1):183–196. doi: 10.1093/nar/10.1.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki Y., Wada C., Yura T. Mini-F plasmid mutants able to replicate in the absence of sigma 32: mutations in the repE coding region producing hyperactive initiator protein. J Bacteriol. 1991 Feb;173(3):1064–1072. doi: 10.1128/jb.173.3.1064-1072.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder P., Churchward G., Caro L. Plasmid pSC101 replication mutants generated by insertion of the transposon Tn1000. J Mol Biol. 1983 Oct 25;170(2):287–303. doi: 10.1016/s0022-2836(83)80149-1. [DOI] [PubMed] [Google Scholar]
- Linder P., Churchward G., Xia G. X., Yu Y. Y., Caro L. An essential replication gene, repA, of plasmid pSC101 is autoregulated. J Mol Biol. 1985 Feb 5;181(3):383–393. doi: 10.1016/0022-2836(85)90227-x. [DOI] [PubMed] [Google Scholar]
- Manen D., Caro L. The replication of plasmid pSC101. Mol Microbiol. 1991 Feb;5(2):233–237. doi: 10.1111/j.1365-2958.1991.tb02103.x. [DOI] [PubMed] [Google Scholar]
- Manen D., Izaurralde E., Churchward G., Caro L. Transcription events in the origin of replication of plasmid pSC101. J Bacteriol. 1989 Dec;171(12):6482–6492. doi: 10.1128/jb.171.12.6482-6492.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manen D., Upegui-Gonzalez L. C., Caro L. Monomers and dimers of the RepA protein in plasmid pSC101 replication: domains in RepA. Proc Natl Acad Sci U S A. 1992 Oct 1;89(19):8923–8927. doi: 10.1073/pnas.89.19.8923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meacock P. A., Cohen S. N. Partitioning of bacterial plasmids during cell division: a cis-acting locus that accomplishes stable plasmid inheritance. Cell. 1980 Jun;20(2):529–542. doi: 10.1016/0092-8674(80)90639-x. [DOI] [PubMed] [Google Scholar]
- Perri S., Helinski D. R., Toukdarian A. Interactions of plasmid-encoded replication initiation proteins with the origin of DNA replication in the broad host range plasmid RK2. J Biol Chem. 1991 Jul 5;266(19):12536–12543. [PubMed] [Google Scholar]
- Prentki P., Krisch H. M. In vitro insertional mutagenesis with a selectable DNA fragment. Gene. 1984 Sep;29(3):303–313. doi: 10.1016/0378-1119(84)90059-3. [DOI] [PubMed] [Google Scholar]
- Stenzel T. T., MacAllister T., Bastia D. Cooperativity at a distance promoted by the combined action of two replication initiator proteins and a DNA bending protein at the replication origin of pSC101. Genes Dev. 1991 Aug;5(8):1453–1463. doi: 10.1101/gad.5.8.1453. [DOI] [PubMed] [Google Scholar]
- Stenzel T. T., Patel P., Bastia D. The integration host factor of Escherichia coli binds to bent DNA at the origin of replication of the plasmid pSC101. Cell. 1987 Jun 5;49(5):709–717. doi: 10.1016/0092-8674(87)90547-2. [DOI] [PubMed] [Google Scholar]
- Studier F. W., Rosenberg A. H., Dunn J. J., Dubendorff J. W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- Sugiura S., Tanaka M., Masamune Y., Yamaguchi K. DNA binding properties of purified replication initiator protein (Rep) encoded by plasmid pSC101. J Biochem. 1990 Mar;107(3):369–376. doi: 10.1093/oxfordjournals.jbchem.a123052. [DOI] [PubMed] [Google Scholar]
- Vocke C., Bastia D. DNA-protein interaction at the origin of DNA replication of the plasmid pSC101. Cell. 1983 Dec;35(2 Pt 1):495–502. doi: 10.1016/0092-8674(83)90183-6. [DOI] [PubMed] [Google Scholar]
- Vocke C., Bastia D. Primary structure of the essential replicon of the plasmid pSC101. Proc Natl Acad Sci U S A. 1983 Nov;80(21):6557–6561. doi: 10.1073/pnas.80.21.6557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner S., Hoskins J., McKenney K. Monomerization of RepA dimers by heat shock proteins activates binding to DNA replication origin. Proc Natl Acad Sci U S A. 1991 Sep 15;88(18):7903–7907. doi: 10.1073/pnas.88.18.7903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia G. X., Manen D., Goebel T., Linder P., Churchward G., Caro L. A copy-number mutant of plasmid pSC101. Mol Microbiol. 1991 Mar;5(3):631–640. doi: 10.1111/j.1365-2958.1991.tb00734.x. [DOI] [PubMed] [Google Scholar]
- Yamaguchi K., Yamaguchi M. The replication origin of pSC101: the nucleotide sequence and replication functions of the ori region. Gene. 1984 Jul-Aug;29(1-2):211–219. doi: 10.1016/0378-1119(84)90181-1. [DOI] [PubMed] [Google Scholar]
- Zuker M. Suboptimal sequence alignment in molecular biology. Alignment with error analysis. J Mol Biol. 1991 Sep 20;221(2):403–420. doi: 10.1016/0022-2836(91)80062-y. [DOI] [PubMed] [Google Scholar]