Abstract
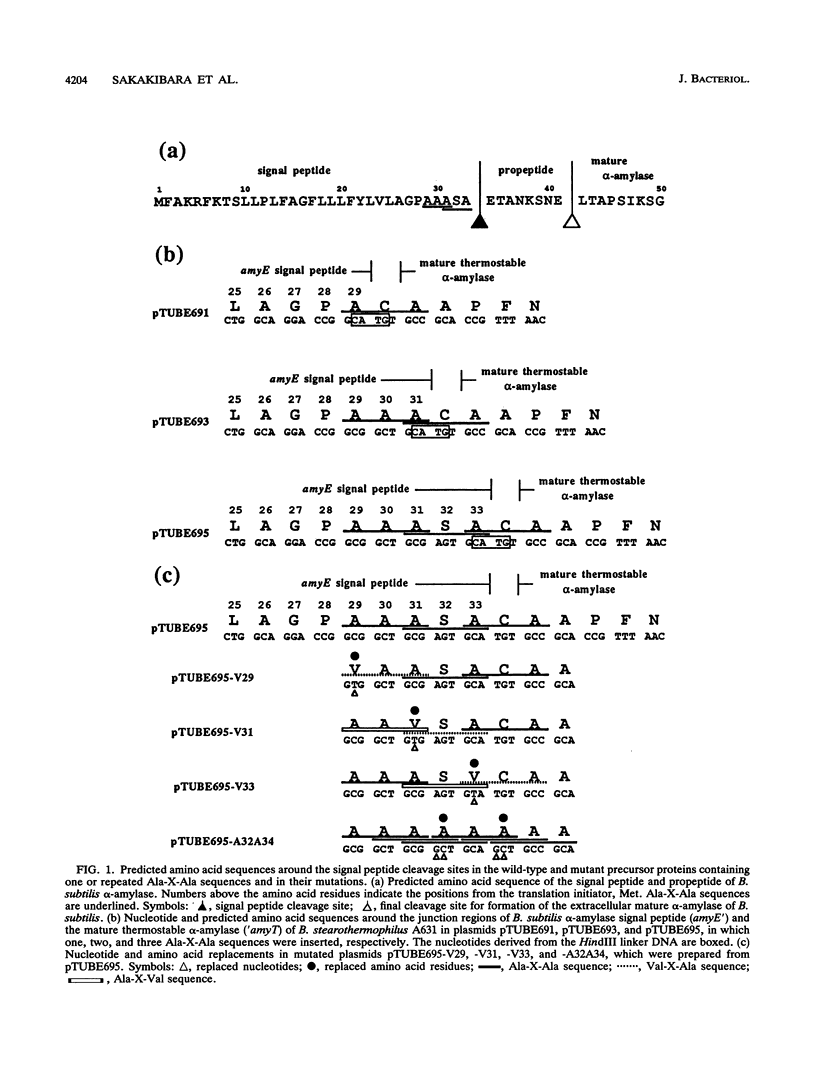
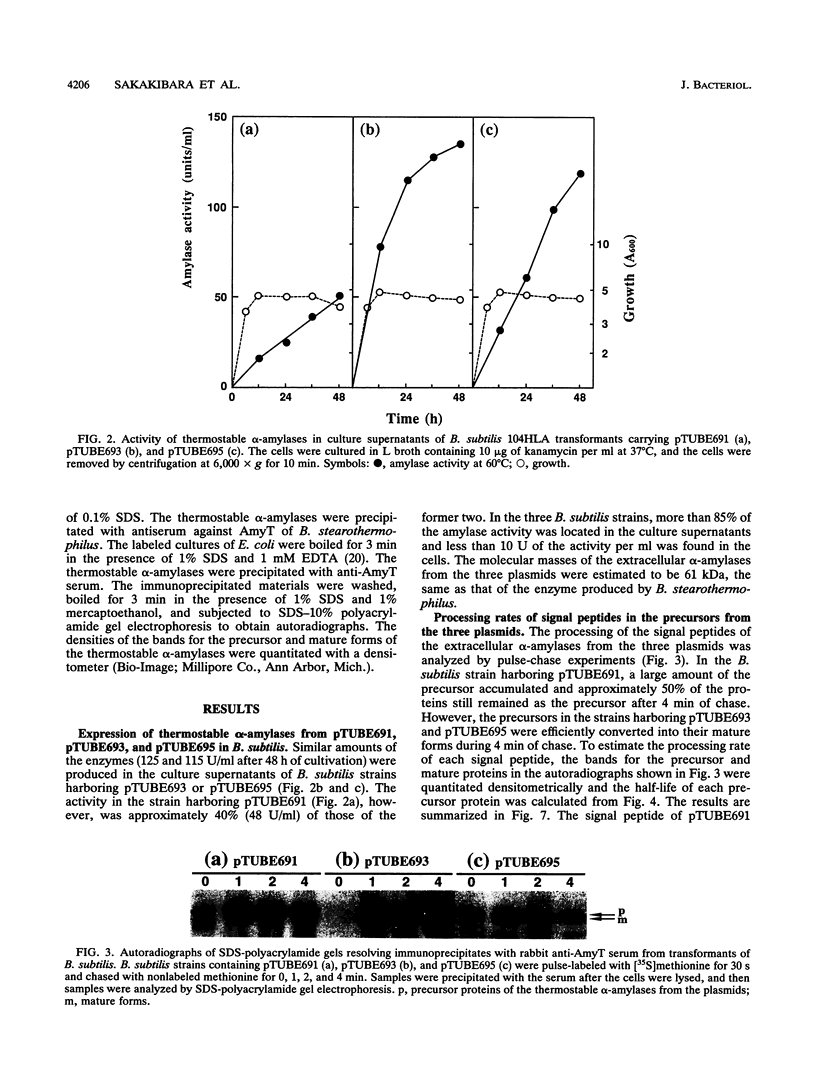
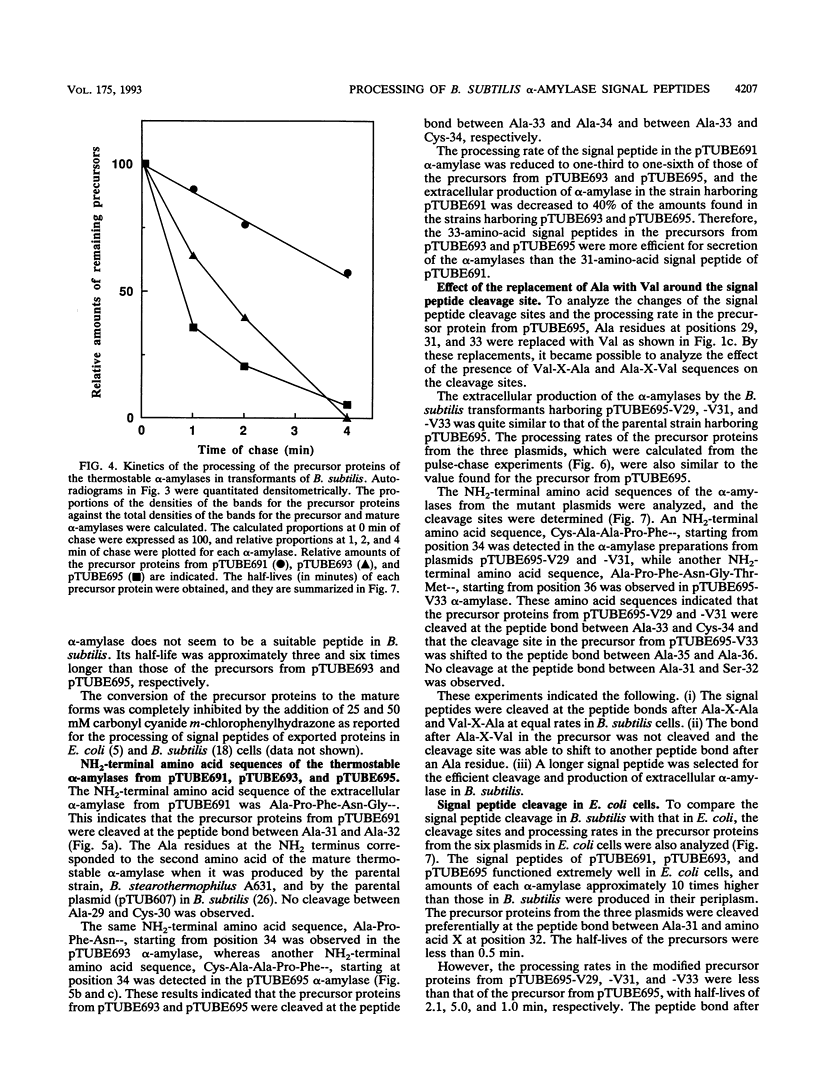
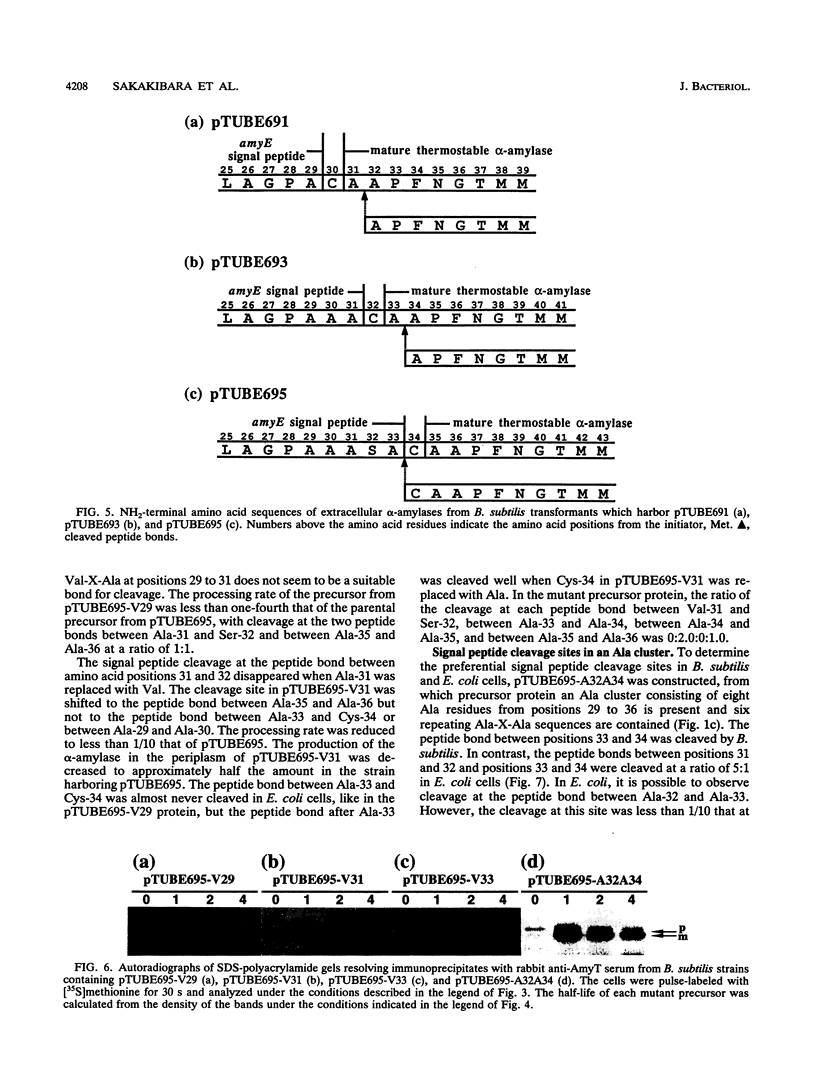
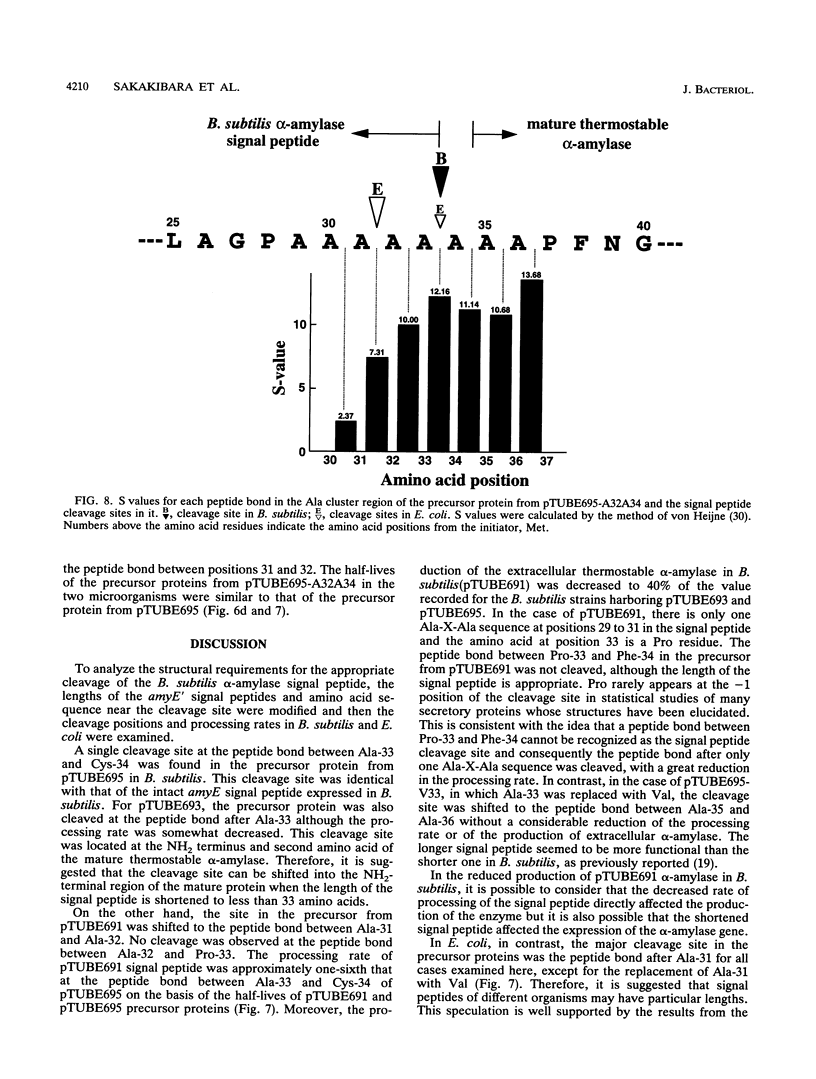
The Bacillus subtilis alpha-amylase signal peptide consists of 33 amino acids from its translation initiation site. To analyze the structural requirements for efficient processing of the signal peptide, single and repeated Ala-X-Ala sequences and their modifications were introduced into B. subtilis alpha-amylase signal peptides of different lengths and the mature thermostable alpha-amylase. Then the cleavage positions and processing rates of the signal peptides were analyzed by the NH2-terminal amino acid sequences of the exported thermostable alpha-amylases and by in vivo pulse-chase experiments. In B. subtilis, the most efficient cleavage site was located at the peptide bond between Ala-33 and amino acid X at position 34, even though Val-X-Ala and six repeating Ala-X-Ala sequences were present around the cleavage site. However, the cleavage site was shifted to the peptide bond between Ala-31 and amino acid X when Ala-33 was deleted, and it was also shifted to Ala-35 and X when Ala-33 was replaced with Val-33. The shorter signal peptide consisting of 31 amino acids reduced the processing rate and alpha-amylase production. In contrast, those signal peptides were cleaved preferentially at the peptide bond between Ala-31 and amino acid X in Escherichia coli. In addition to the presence of an Ala residue at the -1 amino acid position, the length of the signal peptide was another important requirement for efficient processing.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bedouelle H., Bassford P. J., Jr, Fowler A. V., Zabin I., Beckwith J., Hofnung M. Mutations which alter the function of the signal sequence of the maltose binding protein of Escherichia coli. Nature. 1980 May 8;285(5760):78–81. doi: 10.1038/285078a0. [DOI] [PubMed] [Google Scholar]
- Blobel G., Dobberstein B. Transfer of proteins across membranes. I. Presence of proteolytically processed and unprocessed nascent immunoglobulin light chains on membrane-bound ribosomes of murine myeloma. J Cell Biol. 1975 Dec;67(3):835–851. doi: 10.1083/jcb.67.3.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan S. J., Weiss J., Konrad M., White T., Bahl C., Yu S. D., Marks D., Steiner D. F. Biosynthesis and periplasmic segregation of human proinsulin in Escherichia coli. Proc Natl Acad Sci U S A. 1981 Sep;78(9):5401–5405. doi: 10.1073/pnas.78.9.5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S., Cohen S. N. High frequency transformation of Bacillus subtilis protoplasts by plasmid DNA. Mol Gen Genet. 1979 Jan 5;168(1):111–115. doi: 10.1007/BF00267940. [DOI] [PubMed] [Google Scholar]
- Daniels C. J., Bole D. G., Quay S. C., Oxender D. L. Role for membrane potential in the secretion of protein into the periplasm of Escherichia coli. Proc Natl Acad Sci U S A. 1981 Sep;78(9):5396–5400. doi: 10.1073/pnas.78.9.5396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dev I. K., Ray P. H. Signal peptidases and signal peptide hydrolases. J Bioenerg Biomembr. 1990 Jun;22(3):271–290. doi: 10.1007/BF00763168. [DOI] [PubMed] [Google Scholar]
- Gierasch L. M. Signal sequences. Biochemistry. 1989 Feb 7;28(3):923–930. doi: 10.1021/bi00429a001. [DOI] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983 Jun 5;166(4):557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Inouye M., Halegoua S. Secretion and membrane localization of proteins in Escherichia coli. CRC Crit Rev Biochem. 1980;7(4):339–371. doi: 10.3109/10409238009105465. [DOI] [PubMed] [Google Scholar]
- Itoh Y., Kanoh K., Nakamura K., Takase K., Yamane K. Artificial insertion of peptides between signal peptide and mature protein: effect on secretion and processing of hybrid thermostable alpha-amylases in Bacillus subtilis and Escherichia coli cells. J Gen Microbiol. 1990 Aug;136(8):1551–1558. doi: 10.1099/00221287-136-8-1551. [DOI] [PubMed] [Google Scholar]
- Itoh Y., Sumi M., Nakamura K., Yamane K. Processing of an NH2-terminally extended thermostable alpha-amylase by Bacillus subtilis alkaline protease. J Biochem. 1990 Dec;108(6):954–959. doi: 10.1093/oxfordjournals.jbchem.a123320. [DOI] [PubMed] [Google Scholar]
- Kawamura F., Doi R. H. Construction of a Bacillus subtilis double mutant deficient in extracellular alkaline and neutral proteases. J Bacteriol. 1984 Oct;160(1):442–444. doi: 10.1128/jb.160.1.442-444.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi Y., Yoda K., Yamasaki M., Tamura G. The nucleotide sequence of the promoter and the amino-terminal region of alkaline phosphatase structural gene (phoA) of Escherichia coli. Nucleic Acids Res. 1981 Nov 11;9(21):5671–5678. doi: 10.1093/nar/9.21.5671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Matsudaira P. Sequence from picomole quantities of proteins electroblotted onto polyvinylidene difluoride membranes. J Biol Chem. 1987 Jul 25;262(21):10035–10038. [PubMed] [Google Scholar]
- Murén E. M., Randall L. L. Export of alpha-amylase by Bacillus amyloliquefaciens requires proton motive force. J Bacteriol. 1985 Nov;164(2):712–716. doi: 10.1128/jb.164.2.712-716.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura K., Fujita Y., Itoh Y., Yamane K. Modification of length, hydrophobic properties and electric charge of Bacillus subtilis alpha-amylase signal peptide and their different effects on the production of secretory proteins in B. subtilis and Escherichia coli cells. Mol Gen Genet. 1989 Mar;216(1):1–9. doi: 10.1007/BF00332223. [DOI] [PubMed] [Google Scholar]
- Nakamura K., Takamatsu H., Akiyama Y., Ito K., Yamane K. Complementation of the protein transport defect of an Escherichia coli secY mutant (secY24) by Bacillus subtilis secY homologue. FEBS Lett. 1990 Oct 29;273(1-2):75–78. doi: 10.1016/0014-5793(90)81054-r. [DOI] [PubMed] [Google Scholar]
- Perlman D., Halvorson H. O. A putative signal peptidase recognition site and sequence in eukaryotic and prokaryotic signal peptides. J Mol Biol. 1983 Jun 25;167(2):391–409. doi: 10.1016/s0022-2836(83)80341-6. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasamoto H., Nakazawa K., Tsutsumi K., Takase K., Yamane K. Signal peptide of Bacillus subtilis alpha-amylase. J Biochem. 1989 Sep;106(3):376–382. doi: 10.1093/oxfordjournals.jbchem.a122861. [DOI] [PubMed] [Google Scholar]
- Sohma A., Fujita T., Yamane K. Protein processing to form extracellular thermostable alpha-amylases from a gene fused in a Bacillus subtilis secretion vector. J Gen Microbiol. 1987 Nov;133(11):3271–3277. doi: 10.1099/00221287-133-11-3271. [DOI] [PubMed] [Google Scholar]
- Takase K., Mizuno H., Yamane K. NH2-terminal processing of Bacillus subtilis alpha-amylase. J Biol Chem. 1988 Aug 15;263(23):11548–11553. [PubMed] [Google Scholar]
- Watson M. E. Compilation of published signal sequences. Nucleic Acids Res. 1984 Jul 11;12(13):5145–5164. doi: 10.1093/nar/12.13.5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong S. L., Price C. W., Goldfarb D. S., Doi R. H. The subtilisin E gene of Bacillus subtilis is transcribed from a sigma 37 promoter in vivo. Proc Natl Acad Sci U S A. 1984 Feb;81(4):1184–1188. doi: 10.1073/pnas.81.4.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki H., Ohmura K., Nakayama A., Takeichi Y., Otozai K., Yamasaki M., Tamura G., Yamane K. Alpha-amylase genes (amyR2 and amyE+) from an alpha-amylase-hyperproducing Bacillus subtilis strain: molecular cloning and nucleotide sequences. J Bacteriol. 1983 Oct;156(1):327–337. doi: 10.1128/jb.156.1.327-337.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijl J. M., de Jong A., Vehmaanperä J., Venema G., Bron S. Signal peptidase I of Bacillus subtilis: patterns of conserved amino acids in prokaryotic and eukaryotic type I signal peptidases. EMBO J. 1992 Aug;11(8):2819–2828. doi: 10.1002/j.1460-2075.1992.tb05349.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G. Patterns of amino acids near signal-sequence cleavage sites. Eur J Biochem. 1983 Jun 1;133(1):17–21. doi: 10.1111/j.1432-1033.1983.tb07424.x. [DOI] [PubMed] [Google Scholar]