Abstract
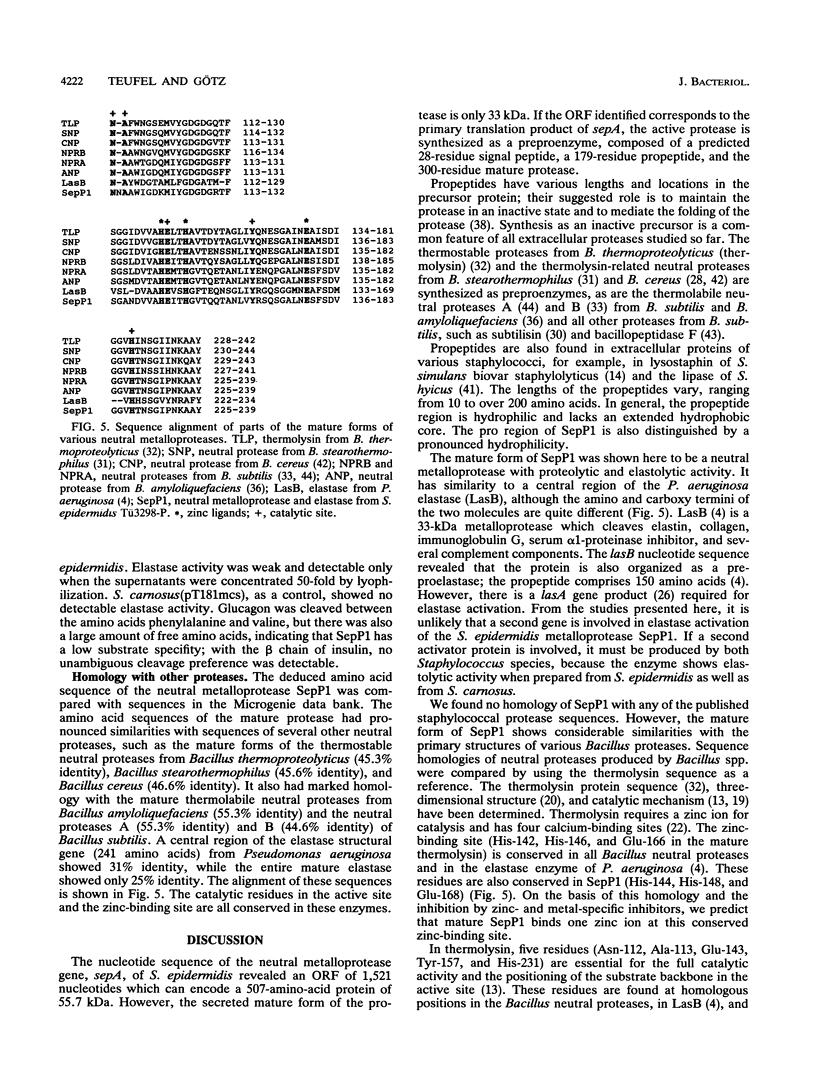
The gene sepA from Staphylococcus epidermidis TU3298-P, encoding the extracellular neutral metalloprotease SepP1, was cloned into pT181mcs. DNA sequencing revealed an open reading frame of 1,521 nucleotides encoding a 507-amino-acid protein with an M(r) of 55,819. The sepA-containing DNA fragment did not hybridize with Staphylococcus hyicus or Staphylococcus carnosus DNA. Expression of sepA in the protease-negative S. carnosus (pT181mcsP1) resulted in overproduction of a 33-kDa protease found in the culture medium. The first 15 N-terminal amino acids of the partially purified protease completely matched the deduced DNA sequence starting at GCA (Ala-208). This finding indicated that SepP1 is synthesized as a preproenzyme with a 28-amino-acid signal peptide, a 179-amino-acid hydrophilic pro region, and a 300-amino-acid extracellular mature form with a calculated M(r) of 32,739. In activity staining, the mature protease prepared from S. carnosus (pT181mcsP1) corresponded to the extracellular S. epidermidis Tü3298-P protease. The partially purified protease had a pH optimum between 5 and 7, and its activity could be inhibited by zinc- and metal-specific inhibitors such as EDTA and 1,10-phenanthroline, indicating that it is a neutral metalloprotease. The protease had a low substrate specificity. Glucagon was cleaved preferentially between aromatic (Phe) and hydrophobic (Val) amino acids. The protease hydrolyzed casein and elastin. The amino acid sequence of the mature form of SepP1 revealed pronounced similarities with the thermolabile and thermostable neutral proteases of various bacilli (44 to 55% identity) and a central part of the mature form of the Pseudomonas aeruginosa elastase (31% identity). From homology comparison with the Bacillus thermoproteolyticus thermolysin, we predict that mature SepP1 binds one zinc ion at a conserved zinc-binding site.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arvidson S., Holme T., Lindholm B. Studies on extracellular proteolytic enzymes from Staphylococcus aureus. I. Purification and characterization of one neutral and one alkaline protease. Biochim Biophys Acta. 1973 Mar 15;302(1):135–148. doi: 10.1016/0005-2744(73)90016-8. [DOI] [PubMed] [Google Scholar]
- Arvidson S. Studies on extracellular proteolytic enzymes from Staphylococcus aureus. II. Isolation and characterization of an EDTA-sensitive protease. Biochim Biophys Acta. 1973 Mar 15;302(1):149–157. doi: 10.1016/0005-2744(73)90017-x. [DOI] [PubMed] [Google Scholar]
- Augustin J., Rosenstein R., Wieland B., Schneider U., Schnell N., Engelke G., Entian K. D., Götz F. Genetic analysis of epidermin biosynthetic genes and epidermin-negative mutants of Staphylococcus epidermidis. Eur J Biochem. 1992 Mar 15;204(3):1149–1154. doi: 10.1111/j.1432-1033.1992.tb16740.x. [DOI] [PubMed] [Google Scholar]
- Bever R. A., Iglewski B. H. Molecular characterization and nucleotide sequence of the Pseudomonas aeruginosa elastase structural gene. J Bacteriol. 1988 Sep;170(9):4309–4314. doi: 10.1128/jb.170.9.4309-4314.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjoörklind A., Jörnvall H. Substrate specificity of three different extracellular proteolytic enzymes from Staphylococcus aureus. Biochim Biophys Acta. 1974 Dec 29;370(2):524–529. doi: 10.1016/0005-2744(74)90113-2. [DOI] [PubMed] [Google Scholar]
- Drapeau G. R., Boily Y., Houmard J. Purification and properties of an extracellular protease of Staphylococcus aureus. J Biol Chem. 1972 Oct 25;247(20):6720–6726. [PubMed] [Google Scholar]
- Drapeau G. R. The primary structure of staphylococcal protease. Can J Biochem. 1978 Jun;56(6):534–544. doi: 10.1139/o78-082. [DOI] [PubMed] [Google Scholar]
- Drapeau G. R. Unusual COOH-terminal structure of staphylococcal protease. J Biol Chem. 1978 Sep 10;253(17):5899–5901. [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Fujishima A., Honda K. Electrochemical photolysis of water at a semiconductor electrode. Nature. 1972 Jul 7;238(5358):37–38. doi: 10.1038/238037a0. [DOI] [PubMed] [Google Scholar]
- Götz F., Zabielski J., Philipson L., Lindberg M. DNA homology between the arsenate resistance plasmid pSX267 from Staphylococcus xylosus and the penicillinase plasmid pI258 from Staphylococcus aureus. Plasmid. 1983 Mar;9(2):126–137. doi: 10.1016/0147-619x(83)90015-x. [DOI] [PubMed] [Google Scholar]
- Hangauer D. G., Monzingo A. F., Matthews B. W. An interactive computer graphics study of thermolysin-catalyzed peptide cleavage and inhibition by N-carboxymethyl dipeptides. Biochemistry. 1984 Nov 20;23(24):5730–5741. doi: 10.1021/bi00319a011. [DOI] [PubMed] [Google Scholar]
- Heinrich P., Rosenstein R., Böhmer M., Sonner P., Götz F. The molecular organization of the lysostaphin gene and its sequences repeated in tandem. Mol Gen Genet. 1987 Oct;209(3):563–569. doi: 10.1007/BF00331163. [DOI] [PubMed] [Google Scholar]
- Khan S. A., Novick R. P. Complete nucleotide sequence of pT181, a tetracycline-resistance plasmid from Staphylococcus aureus. Plasmid. 1983 Nov;10(3):251–259. doi: 10.1016/0147-619x(83)90039-2. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Matsudaira P. Sequence from picomole quantities of proteins electroblotted onto polyvinylidene difluoride membranes. J Biol Chem. 1987 Jul 25;262(21):10035–10038. [PubMed] [Google Scholar]
- Recsei P. A., Gruss A. D., Novick R. P. Cloning, sequence, and expression of the lysostaphin gene from Staphylococcus simulans. Proc Natl Acad Sci U S A. 1987 Mar;84(5):1127–1131. doi: 10.1073/pnas.84.5.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche R. S., Voordouw G. The structural and functional roles of metal ions in thermolysin. CRC Crit Rev Biochem. 1978;5(1):1–23. doi: 10.3109/10409237809177138. [DOI] [PubMed] [Google Scholar]
- Rosenstein R., Peschel A., Wieland B., Götz F. Expression and regulation of the antimonite, arsenite, and arsenate resistance operon of Staphylococcus xylosus plasmid pSX267. J Bacteriol. 1992 Jun;174(11):3676–3683. doi: 10.1128/jb.174.11.3676-3683.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rydén A. C., Rydén L., Philipson L. Isolation and properties of a staphylococcal protease, preferentially cleaving glutamoyl-peptide bonds. Eur J Biochem. 1974 May 2;44(1):105–114. doi: 10.1111/j.1432-1033.1974.tb03462.x. [DOI] [PubMed] [Google Scholar]
- SCHINDLER C. A., SCHUHARDT V. T. PURIFICATION AND PROPERTIES OF LYSOSTAPHIN--A LYTIC AGENT FOR STAPHYLOCOCCUS AUREUS. Biochim Biophys Acta. 1965 Feb 15;97:242–250. doi: 10.1016/0304-4165(65)90088-7. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schad P. A., Iglewski B. H. Nucleotide sequence and expression in Escherichia coli of the Pseudomonas aeruginosa lasA gene. J Bacteriol. 1988 Jun;170(6):2784–2789. doi: 10.1128/jb.170.6.2784-2789.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Stahl M. L., Ferrari E. Replacement of the Bacillus subtilis subtilisin structural gene with an In vitro-derived deletion mutation. J Bacteriol. 1984 May;158(2):411–418. doi: 10.1128/jb.158.2.411-418.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi M., Imanaka T., Aiba S. Nucleotide sequence and promoter region for the neutral protease gene from Bacillus stearothermophilus. J Bacteriol. 1985 Sep;163(3):824–831. doi: 10.1128/jb.163.3.824-831.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran L., Wu X. C., Wong S. L. Cloning and expression of a novel protease gene encoding an extracellular neutral protease from Bacillus subtilis. J Bacteriol. 1991 Oct;173(20):6364–6372. doi: 10.1128/jb.173.20.6364-6372.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trayer H. R., Buckley C. E., 3rd Molecular properties of lysostaphin, a bacteriolytic agent specific for Staphylococcus aureus. J Biol Chem. 1970 Sep 25;245(18):4842–4846. [PubMed] [Google Scholar]
- Twining S. S. Fluorescein isothiocyanate-labeled casein assay for proteolytic enzymes. Anal Biochem. 1984 Nov 15;143(1):30–34. doi: 10.1016/0003-2697(84)90553-0. [DOI] [PubMed] [Google Scholar]
- Vasantha N., Thompson L. D., Rhodes C., Banner C., Nagle J., Filpula D. Genes for alkaline protease and neutral protease from Bacillus amyloliquefaciens contain a large open reading frame between the regions coding for signal sequence and mature protein. J Bacteriol. 1984 Sep;159(3):811–819. doi: 10.1128/jb.159.3.811-819.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wandersman C. Secretion, processing and activation of bacterial extracellular proteases. Mol Microbiol. 1989 Dec;3(12):1825–1831. doi: 10.1111/j.1365-2958.1989.tb00169.x. [DOI] [PubMed] [Google Scholar]
- Watson M. E. Compilation of published signal sequences. Nucleic Acids Res. 1984 Jul 11;12(13):5145–5164. doi: 10.1093/nar/12.13.5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver L. H., Kester W. R., Matthews B. W. A crystallographic study of the complex of phosphoramidon with thermolysin. A model for the presumed catalytic transition state and for the binding of extended substances. J Mol Biol. 1977 Jul;114(1):119–132. doi: 10.1016/0022-2836(77)90286-8. [DOI] [PubMed] [Google Scholar]
- Wetmore D. R., Wong S. L., Roche R. S. The role of the pro-sequence in the processing and secretion of the thermolysin-like neutral protease from Bacillus cereus. Mol Microbiol. 1992 Jun;6(12):1593–1604. doi: 10.1111/j.1365-2958.1992.tb00884.x. [DOI] [PubMed] [Google Scholar]
- Yang M. Y., Ferrari E., Henner D. J. Cloning of the neutral protease gene of Bacillus subtilis and the use of the cloned gene to create an in vitro-derived deletion mutation. J Bacteriol. 1984 Oct;160(1):15–21. doi: 10.1128/jb.160.1.15-21.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 1986 Jun 11;14(11):4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]



