Abstract
Asexual spores (conidia) are the infectious propagules of many pathogenic fungi, and the capacity to sense the host environment and trigger conidial germination is a key pathogenicity determinant. Germination of conidia requires the de novo establishment of a polarised growth axis and consequent germ tube extension. The molecular mechanisms that control polarisation during germination are poorly understood. In the dimorphic human pathogenic fungus Penicillium marneffei, conidia germinate to produce one of two cell types that have very different fates in response to an environmental cue. At 25 °C, conidia germinate to produce the saprophytic cell type, septate, multinucleate hyphae that have the capacity to undergo asexual development. At 37 °C, conidia germinate to produce the pathogenic cell type, arthroconidiating hyphae that liberate uninucleate yeast cells. This study shows that the p21-activated kinase pakA is an essential component of the polarity establishment machinery during conidial germination and polarised growth of yeast cells at 37 °C but is not required for germination or polarised growth at 25 °C. Analysis shows that the heterotrimeric G protein α subunit GasC and the CDC42 orthologue CflA lie upstream of PakA for germination at both temperatures, while the Ras orthologue RasA only functions at 25 °C. These findings suggest that although some proteins that regulate the establishment of polarised growth in budding yeast are conserved in filamentous fungi, the circuitry and downstream effectors are differentially regulated to give rise to distinct cell types.
Author Summary
Many fungal infections are initiated by the entry of dormant fungal spores into their host. Once inside the host these dormant spores must reactivate (germinate) for the infection to proceed. Productive infections necessitate that the fungus grow and divide within the host, which makes understanding the mechanisms that control germination crucial to developing preventative or prophylactic treatments for fungal infections. The molecular mechanisms that control spore germination are poorly understood and studies of the opportunistic fungal pathogen Penicillium marneffei have shown that a group of highly conserved signalling and cell polarity factors, known as small GTPases, play important roles in germination and other aspects of morphogenesis. In this study we have shown that a downstream target of these small GTPases, a p21-activated kinase plays a crucial role in germination at the host temperature of 37 °C but not at 25 °C. This is the first component of germination, which shows temperature-dependent effects and provides insights into the different mechanisms used by fungal pathogens to infect their host or to grow saprophytically in non-host environments.
Introduction
The generation of an axis of cell polarity is central to the activity of many cells and the establishment of a wide variety of cell morphologies. It relies on the ability to mark different cellular regions by specific protein localisation. The establishment of polarised growth requires selection of a site to which proteins and components of the cytoskeleton are recruited. Growth is then directed specifically to this site via targeted cellular trafficking and concomitant cell wall deposition. Fungi are small eukaryotes that exhibit highly polarised growth patterns and provide excellent models for the study of the molecular mechanisms underlying cell polarity. Saccharomyces cerevisiae establishes a polarised axis of growth during the processes of budding cell division, schmoo formation during mating, and pseudohyphal growth. Under conditions of nitrogen starvation, S. cerevisiae diploid cells undergo pseudohyphal growth, a morphological switch that involves changes in cell shape and division [1]. Pseudohyphal growth requires the initiation of polarised growth for cellular elongation and is under the control of two signaling pathways: a cyclic adenosine monophosphate (cAMP)/protein kinase A (PKA) pathway and a mitogen-activated protein kinase (MAPK) cascade. The cAMP/PKA pathway is activated by a glucose/sucrose sensitive receptor Gpr1p, which activates the G protein Gpa2p (α subunit), which in turn is inhibited by the novel kelch-Gβ subunits Gpb1/2p, a third subunit Gpg1p, and a negative regulator Rgs2p [2–8]. The Gpa2-Gpb1/2 complex regulates the cAMP/PKA pathway directly in association with the Ras2p GTPase and the RasGAP neurofibromin homologues Ira1/2p [6–7, 9]. Ras2p can also activate the MAPK cascade by activating the guanine nucleotide exchange factor Cdc24p, which catalyzes the guanosine diphosphate (GDP) to guanosine triphosphate (GTP) exchange of the Rho GTPase Cdc42p [1,10,11]. GTP-bound Cdc42p is required to initiate actin polarisation and recruits and activates additional proteins required for polarised growth such as septins, myosins, and the p21-activated kinase (PAK) Ste20p (reviewed in [12]). In turn, Ste20p activates the MAPK cascade by phosphorylating Ste11p (MAPKKK); Ste11p phosphorylates Ste7p (MAPKK), which then activates the Kss1p MAPK [13–15].
Filamentous fungi exhibit a highly polarised axis during vegetative hyphal growth, asexual (conidiation) and sexual (mating) development, and initiation of polarised filamentous growth during the germination of asexual (conidia) and sexual (ascospores) spores. Spherical conidia germinate under favorable environmental conditions by initially growing isotropically. This growth is followed by the establishment of a de novo axis of polarised growth to allow a germ tube to emerge [16–18]. Conidial germination is a central aspect of fungal cell propagation, initiating the formation of the extensive radiating hyphal network necessary for colonization of substrates from a dormant conidium. Conidia are also the infectious propagules of many pathogenic fungi, and germination of conidia in the lung or leaves of potential hosts is likely to be a key pathogenicity determinant. Studies in the filamentous fungus Aspergillus nidulans have shown that the cAMP-PKA and Ras pathways play a role [18–20]. However, the molecular mechanisms governing germination of fungal conidia are not well understood in any system.
Penicillium marneffei is an opportunistic human pathogen with a thermally regulated dimorphic switch. At 25 °C, in the saprophytic growth phase, conidia germinate to produce highly polarised, septate, branched, multinucleate hyphae. Conidia can also germinate at 37 °C to produce polarised arthroconidiating hyphae, in which nuclear division and septation are coupled, double septa are laid down, and fragmentation occurs along this plane to liberate uninucleate yeast cells that consequently divide by fission [21]. The yeast cells are the pathogenic growth form and multiple yeast cells are observed in the pulmonary alveolar macrophages of infected individuals. P. marneffei infection is thought to occur through inhalation of conidia, which bind to the laminin in the broncholalvelolar epithelia. Conidia are then ingested by pulmonary alveolar macrophages and germinate, generating the uninucleate yeast cells [21]. Therefore in P. marneffei, conidial germination can lead to two very different morphological programs in response to different temperatures.
Some of the core components regulating polarised growth establishment in S. cerevisiae are conserved during polarity establishment in germinating conidia of filamentous fungi. The P. marneffei Gpa2p homologue, encoded by gasC, is required during conidial germination to produce hyphae. Deletion of gasC results in delayed germination, whereas expression of a dominant activating allele shows a significantly accelerated germination rate [22]. Likewise, the P. marneffei Ras homologue, RasA, is required for conidial germination where expression of either a dominant negative or activated allele results in a germination delay and conidia with abnormal isotropic growth [23]. Expression of either a dominant negative or constitutively active allele of the P. marneffei CDC42 homologue (cflA) results in a decrease or increase in the rate of germination at 25 °C, respectively [24]. The role of GasC, RasA, and CflA during conidial germination in P. marneffei suggests that the core components regulating polarised growth establishment in S. cerevisiae may be conserved during polarity establishment in germinating conidia of filamentous fungi. To investigate if any of the downstream effectors of this core pathway are also conserved in function, a PAK STE20 homologue (designated pakA) was identified and deleted in P. marneffei. Characterization of pakA in P. marneffei has shown that this gene is essential for conidial germination at 37 °C and for polarised growth of yeast cells and is downstream of both a heterotrimeric G protein and Cdc42 pathway. In contrast, PakA plays only a minor role during germination of conidia at 25 °C and is not required for polarised growth of hyphae. Germination in this case is controlled by a heterotrimeric G protein–Ras-Cdc42 pathway. These data suggest that although some proteins that regulate the establishment of polarised growth in budding yeast and filamentous fungi may be conserved, the downstream effectors are likely to be different or regulated differently to give rise to the distinct cell types in these two modes of growth.
Results
Cloning the STE20 p21-Activated Kinase Orthologue from P. marneffei
An A. nidulans sequence was identified from the genome sequence (http://www.broad.mit.edu/annotation/genome/aspergillus_group/MultiHome.html) with strong homology to Candida albicans Cst20p (67% identity, 81% similarity) and other PAKs, including S. cerevisiae Ste20p (71% identity, 81% similarity, accession number AAA35039). Primers were designed to amplify the sequence encoding the conserved kinase domain and a PCR product was generated using A. nidulans genomic DNA. The A. nidulans STE20 homologous sequence was used to screen a P. marneffei genomic library at low stringency. Five positive clones were identified, which fell into two classes based on restriction enzyme digestion patterns. A 6.4 kb NotI/BglII hybridizing fragment from one of these classes was subcloned into NotI/BamHI digested pBluescript II SK+ (pKB5751). Sequencing revealed strong sequence homology to STE20-like PAKs from Magnaporthe grisea (78% identity, 89% similarity, accession number AAP93639), Ustilago maydis (74% identity, 83% similarity, accession number AAM97788), and S. cerevisiae (72% identity, 83% similarity). The gene within this clone was subsequently named pakA. The pakA open reading frame spans 2407 bp and contains seven exons and six introns. The predicted protein is 642 amino acids in length and contains a Cdc42/Rac interactive binding (CRIB) domain at positions 98–115, a predicted kinase domain at 361–612, and a Gβ binding domain at 619–629. Preliminary analysis of the second class of clones revealed that the gene within these clones has strong sequence homology to CLA4-like PAKs.
RNA was isolated from vegetative hyphae grown for 2 d in liquid medium at 25 °C, asexual development (conidiation) cultures grown for 4 d on solid medium at 25 °C, and yeast cells grown for 8 d in liquid medium at 37 °C. A pakA transcript was detected under all conditions. The amount of pakA transcript was approximately equivalent under all conditions when compared with the histone H3 control (unpublished data).
Deletion of pakA in P. marneffei
A pakA construct, which deleted from −425 to +2030 of pakA, was linearised and transformed into P. marneffei strain SPM4 (niaD1 pyrG1) and pyrG+ transformants selected. PyrG+ transformants were screened by genomic Southern blotting and one strain was isolated that possessed a restriction pattern consistent with replacement of pakA by pyrG at the genomic locus. The deletion strain was plated on medium containing 5-fluoroorotic acid (5-FOA) to generate a ΔpakA pyrG strain (Materials and Methods). This strain was cotransformed with plasmids containing pakA+ and pyrG+ genes and co-transformants confirmed by Southern blot analysis. The transformants contained from 1–7 copies of pakA.
In S. cerevisiae, the H345G mutation in the conserved CRIB domain of Ste20p results in a mutant protein that shows reduced interaction with the upstream activator Cdc42p (GTPase) in a two-hybrid assay and loss of correct localisation to sites of polarised growth [25]. The phenotype of the STE20H345G strain is almost equivalent to the null, indicating that the interaction of Cdc42p and Ste20p is essential for Ste20p function [25]. The equivalent mutation was generated in P. marneffei pakA (pakAH108G) by inverse PCR (Materials and Methods). The pakAH108G construct was co-transformed with pyrG+ into the ΔpakA pyrG− strain and transformants selected for PyrG+. Co-transformation was confirmed by Southern blot analysis of genomic DNA and four representative transformants with copy numbers ranging from 3 to 12 were selected for further analysis.
PakA Is Localised to Sites of Polarised Growth
The gfp::pakA and gfp::pakAH108G fusion constructs were generated and co-transformed with the pyrG+ gene into the ΔpakA pyrG strain to investigate the localisation of PakA and to assess whether the pakAH108G mutation affects PakA localisation. Transformants were selected for PyrG+ and confirmed by Southern blot analysis of genomic DNA. Four transformants of each genotype were selected for further analysis and had copy numbers ranging from 4 to 9 for gfp::pakA and from 2 to 20 for gfp::pakAH108G. The gfp::pakA and gfp::pakAH108G strains were grown on agar-coated slides for 2 and 4 d at 25 °C and 37 °C, respectively. At 25 °C, the GFP::PakA fusion protein was concentrated at the hyphal apex (Figure 1A) and localised as spots along the subapical cells of the hyphae (Figure 1B). In contrast, the GFP::PakAH108G fusion protein was visible as diffuse fluorescence in the cytoplasm but not concentrated at the hyphal apex or as spots along the hyphae (Figure 1A and 1B). At 37 °C, the GFP::PakA fusion protein was concentrated at the apex of arthroconidiating hyphae, although this fluorescence was less intense than that visualized in the same transformants at 25 °C. The GFP::PakAH108G fusion protein was not observed at the apex of arthroconidiating hyphae (unpublished data).
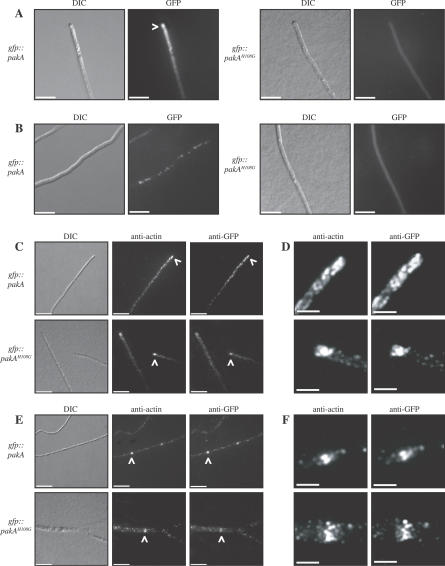
Figure 1. PakA Co-Localises with Actin at Sites of Polarised Growth.
Localisation of PakA in live cells (A, B) and in immunofluorescently labeled fixed cells (C–F) at 25 °C. (A) The GFP::PakA fusion protein is localised to the hyphal apex at 25 °C (white arrowhead). In contrast, the GFP::PakAH108G fusion protein is visible throughout the cytoplasm. (B) In subapical hyphal cells, the GFP::PakA fusion protein is observed as spots along subapical hyphal cells. This localisation is not observed in the gfp::pakAH108G strains. (C) The PakA and PakAH108G GFP fusion proteins co-localised with actin at the hyphal tip (indicated by white arrowheads). (D) Enlargement of the stained region indicated by arrowheads in C. (E) At 25 °C the PakA and PakAH108G GFP fusion proteins also co-localise with actin at nascent septation sites (indicated by arrowheads). (F) Enlargement of the stained region indicated by arrowheads in E. Images were captured using DIC or under epifluorescence to detect GFP, anti-actin, or anti-GFP. Scale bars, 20 μm.
To investigate whether the GFP::PakA fusion protein co-localises with actin at the hyphal apex and at nascent septation sites, immunostaining using mouse anti-actin and rabbit anti-GFP antibodies was performed on two of the gfp::pakA and two of the gfp::pakAH108G strains at both 25 °C and 37 °C. At 25 °C, actin is localised as cortical actin spots along the hyphae and is concentrated at nascent septation sites and the hyphal apex (Figure 1C–1F). The GFP::PakA fusion protein co-localised with actin at all of these locations (Figure 1C–1F). The GFP::PakAH108G fusion protein also co-localised with actin at cortical actin patches, nascent septation sites, and the hyphal apex, in addition to showing diffuse staining throughout the cytoplasm (Figure 1C–1F).
At 37 °C, actin is also localised as cortical patches along the arthroconidiating hyphae, concentrated at nascent septation sites and the apex of arthroconidiating hyphae (Figure S1). The GFP::PakA and GFP::PakAH108G fusion proteins co-localised with actin at all of these sites (Figure S1).
The Role of pakA during Asexual Development and Hyphal Growth at 25 °C
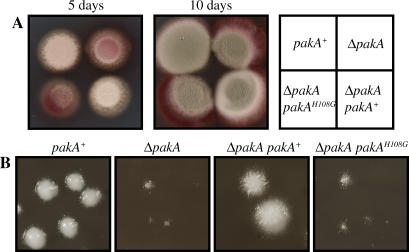
At 25 °C, P. marneffei colonies are comprised of highly polarised vegetative hyphae growing on the agar surface and bearing asexual structures (conidiophores) that appear green due to the presence of pigmented asexual spores (conidia) on the conidiophores. After 5 d growth at 25 °C, surface hyphae are visible in the wild-type strain (Figure 2A). The ΔpakA pakA+ strains appeared wild-type after 5 d at 25° C, whereas both the ΔpakA and ΔpakA pakAH108G strains showed a reduction in growth (Figure 2A). Despite the initial reduction in growth, after 10 d all strains were producing conidia. P. marneffei colonies are yeast-like at 37 °C and ΔpakA pakA+ strains were indistinguishable from the wild type when grown for 4 d at 37 °C (Figure 2B). In contrast, the ΔpakA and ΔpakA pakAH108G strains displayed reduced growth at 37 °C (Figure 2B).
Figure 2. Vegetative Growth Is Reduced in Mutant pakA Strains.
(A) Hyphal growth of the wild-type (pakA+), ΔpakA, ΔpakA pakAH108G, and ΔpakA pakA+ strains after 5 and 10 d incubation at 25 °C. The ΔpakA and ΔpakA pakAH108G strains show reduced growth as evidenced by the red colony appearance due to significantly reduced aerial hyphal growth after 5 d. After 10 d, all strains are conidiating. (B) Yeast colonies of the wild-type (pakA+), ΔpakA, ΔpakA pakAH108G, and ΔpakA pakA+ strains after 5 d at 37 °C. The ΔpakA and ΔpakA pakAH108G strains show reduced growth at 37 °C.
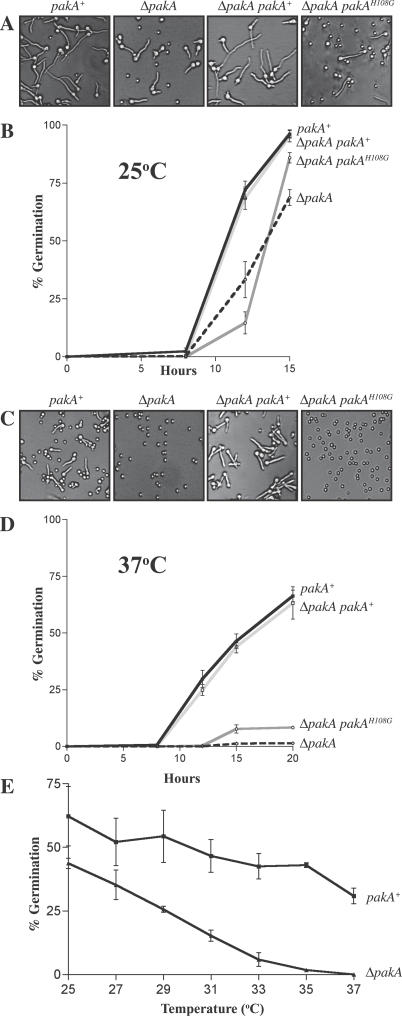
Both the ΔpakA and ΔpakA pakAH108G strains exhibit a delay in growth after 5 d at 25 °C (Figure 2A). One explanation for this difference could be a delay in the germination of conidia. The kinetics of germination were measured at 25 °C in the wild-type (pakA+), ΔpakA, ΔpakA pakA+, and two ΔpakA pakAH108G strains by counting the number of ungerminated versus germinated conidia (conidia with a visible germ tube) in a population of 100 in three independent experiments after 8, 12, or 15 h incubation in liquid media (Table S1). The complemented ΔpakA strain (ΔpakA pakA+) is indistinguishable from wild type (Figure 3A and 3B). The ΔpakA and ΔpakA pakAH108G strains show a minor delay in germination at 25 °C (Figure 3A and 3B).
Figure 3. Mutation of pakA Affects Conidial Germination at 25 °C and 37 °C.
(A) Conidia from the wild-type (pakA+), ΔpakA, ΔpakA pakA+, and ΔpakA pakAH108G strains were grown in liquid medium for 12 h at 25 °C (A) or 37 °C (C). The percentage of germinated conidia was measured after 8, 12, or 15 h incubation at 25 °C (B) or after 8, 12, 15, and 20 h at 37 °C (D). One representative strain of each genotype is shown. Three independent experiments were performed and standard error of the mean bars are shown. (A) Compared with wild-type and the complemented ΔpakA pakA+ strain, the ΔpakA and ΔpakA pakAH108G strains have a larger number of ungerminated conidia visible after 12 h incubation at 25 °C. (B) In contrast to the wild-type (pakA+) and ΔpakA pakA+ strain, the ΔpakA and ΔpakA pakAH108G strains show slightly delayed germination at 25 °C. (C) Unlike wild-type (pakA+) and the complemented ΔpakA pakA+ strain, conidia of the ΔpakA and ΔpakA pakAH108G strains remain predominately ungerminated after 12 h incubation at 37 °C. (D) The ΔpakA and ΔpakA pakAH108G strains show a severe germination defect at 37 °C, with only a small proportion of conidia germinating. (E) Conidia of the wild-type and ΔpakA strain were incubated in liquid medium at 2 °C increments from 25 °C to 37 °C. The percentage of germination was assessed after 20 h. The ΔpakA strain shows a gradual decrease in the percentage of germinated conidia as the temperature increases.
To investigate if the deletion of pakA or the pakAH108G mutation results in aberrant hyphal morphology or asexual development at 25 °C the wild-type, ΔpakA, ΔpakA pakA+, and ΔpakA pakAH108G strains were grown on agar-coated slides for 2 or 4 d at 25 °C, stained with calcofluor (to observe cell walls) or 4′6-diamidino-2-phenylindole (DAPI; to observe nuclei), and examined microscopically. After 2 d at 25 °C, wild-type P. marneffei grows as septate, branched hyphae of which subapical cells are predominately uninucleate and apical cells are multinucleate. The ΔpakA, ΔpakA pakA+, and ΔpakA pakAH108G strains were indistinguishable from wild type after 2 d, with normal morphology, septation, branching, and nuclear index. After 4 d at 25 °C, wild-type P. marneffei begins to undergo asexual development, with the production of a specialized stalk from which differentiated cells are produced sequentially in a budding fashion: metulae bud from the stalk, phialides bud from metulae, and uninucleate conidia bud from phialides. The ΔpakA, ΔpakA pakA+, and ΔpakA pakAH108G strains produced conidiophores with wild-type morphology (unpublished data).
pakA Is Essential for Conidial Germination and Correct Yeast Cell Morphogenesis at 37 °C
In contrast to the wild-type and ΔpakA pakA+ strains, the ΔpakA and ΔpakA pakAH108G strains displayed reduced growth rates at 37 °C (Figure 2B). To assess if the basis of this difference is because pakA is required during the germination of conidia or during yeast morphogenesis and growth at 37 °C, the wild-type (pakA+), ΔpakA, ΔpakA pakA+, and ΔpakA pakAH108G strains were inoculated on agar-coated slides and incubated for 4 d at 37 °C. It was immediately apparent that the ΔpakA and ΔpakA pakAH108G strains possessed a severe germination defect and almost all of the conidia remained ungerminated. The germination kinetics were measured by counting the number of ungerminated versus germinated conidia (conidia with a visible germ tube) in a population of 100 in three independent experiments after 8, 12, 15, or 20 h in liquid medium (Table S2). Germination is slower and germlings appear fatter at 37 °C compared with 25 °C (Figure 3A and 3C). In contrast to wild-type and the ΔpakA pakA+ strains, both the ΔpakA strain and the ΔpakA pakAH108G strains showed a severe defect in germination (Figure 3C and 3D). Despite the majority of conidia remaining ungerminated in the ΔpakA and ΔpakA pakAH108G strains, a small proportion do germinate, and it is presumably these cells that go on to establish the colony.
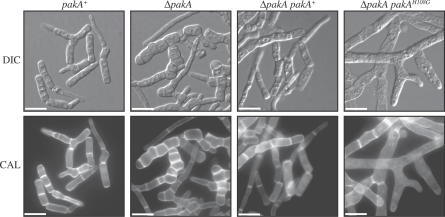
Growth of the pakA strains in liquid medium showed that the small proportion of conidia that germinate go on to form arthroconidiating hyphae that fragment at septation sites to liberate uninucleate yeast cells, just like the wild type. The wild-type (pakA+), ΔpakA, ΔpakA pakA+, and ΔpakA pakAH108G strains were grown in liquid brain heart infusion (BHI) for 6 d at 37 °C and cells were stained with calcofluor or DAPI and observed microscopically (Figure 4). In wild type, the culture consists of a mixture of fragmented arthroconidiating hyphae and uninucleate yeast cells. The ΔpakA strain produced swollen arthroconidiating hyphae and yeast cells with increased septation (Figure 4) but normal nuclear index. These defects were complemented when the strain was transformed with pakA+. The ΔpakA pakAH108G strains also produced swollen arthroconidiating hyphae and yeast cells, but the phenotype was more severe than the ΔpakA strain, with very few yeast cells produced and—in contrast to the ΔpakA strain and wild type—a decrease in the septation index (Figure 4). Therefore, the pakAH108G allele has an inhibitory activity on septation.
Figure 4. pakA Strains Show Aberrant Yeast Morphogenesis at 37 °C.
The wild-type (pakA+), ΔpakA, ΔpakA pakA+, and ΔpakA pakAH108G strains were grown in liquid BHI medium at 37 °C for 6 d. Wild type (pakA+) produces arthroconidiating hyphae and a mixture of fragmented and separated yeast cells. The ΔpakA pakA+ strains are indistinguishable from wild type. The ΔpakA mutant produces swollen arthroconidiating hyphae with increased septation. Fewer yeast cells, which are also swollen and highly septate, are produced in the ΔpakA mutant. The ΔpakA pakAH108G strains produce swollen arthroconidiating hyphae that have reduced septation, and there are reduced numbers of yeast cells. Images were captured using DIC or with epifluorescence to observe calcofluor stained cell walls (CAL). Scale bars, 20 μm.
PakA Is Required for Conidial Germination In Vivo
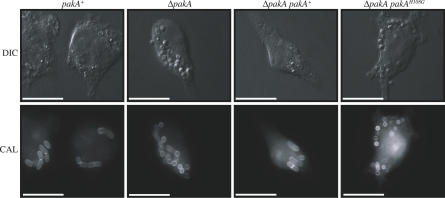
P. marneffei infection occurs through the inhalation of conidia. The conidia are ingested by host pulmonary alveolar macrophages where they germinate into unicellular yeast cells that divide by fission. Multiple yeast cells are seen in the pulmonary alveolar macrophages and peripheral blood mononuclear cells of infected individuals [21]. To investigate if perturbing conidial germination will influence pathogenicity, the ability of the mutant pakA strains to germinate into pathogenic yeast cells was investigated. LPS activated J774 murine macrophages were infected with no conidia or with conidia of the wild-type (pakA+), ΔpakA, ΔpakA pakAH108G, or ΔpakA pakA+ strains (Materials and Methods). After 24 h, numerous yeast cells dividing by fission were observed in macrophages infected with wild-type (pakA+) or ΔpakA pakA+ conidia (Figure 5). Only 16.3 ± 2.50% or 21.5 ± 2.07% of wild-type (pakA+) or ΔpakA pakA+ conidia, respectively, remained ungerminated in infected macrophages. In contrast, conidia of the ΔpakA or ΔpakA pakAH108G strains remained predominately ungerminated in infected macrophages (Figure 5). 60.0 ± 3.80% of ΔpakA conidia and 66.1 ± 7.16% of ΔpakA pakAH108G conidia remained ungerminated in macrophages 24 h post infection. This suggests that PakA is required for conidial germination during infection of host macrophages.
Figure 5. PakA Is Required for Conidial Germination In Vivo.
LPS activated J774 murine macrophages infected with conidial suspensions of the wild-type (pakA+), ΔpakA, ΔpakA pakAH108G, and ΔpakA pakA+ strains. After 24 h, numerous yeast cells dividing by fission were observed in macrophages infected with wild-type (pakA+) or ΔpakA pakA+ conidia. In contrast, conidia of both the ΔpakA and ΔpakA pakAH108G strains remain predominately ungerminated in infected macrophages. Images were captured using DIC or with epifluorescence to observe calcofluor stained fungal cell walls (CAL). Scale bars, 20 μm.
Differential Regulation of Conidial Germination at 25 °C and 37 °C
The temperature-dependent regulation of conidial germination could be the result of the presence of a temperature-specific factor on which PakA depends or the result of a change in the thermostability of a complex in which PakA operates. To distinguish between these two possibilities, wild-type and ΔpakA conidia were incubated in liquid medium at different temperatures ranging from 25 °C to 37 °C, in 2 °C increments, and germination rates were measured (Materials and Methods). In contrast to the wild type, the ΔpakA strain showed a gradual decrease in the percentage of germination as the temperature increased, indicating that there is no critical temperature during the switch (Figure 3E). This result supports the latter hypothesis and suggests PakA-dependent thermosensitivity. To investigate if ΔpakA conidia at 37 °C are waiting for the signal to germinate or have aborted, ΔpakA conidia were incubated at 37 °C for 20 h, then at 25 °C for 20 h. The majority of conidia germinated upon switching to 25 °C, indicating that after 20 h incubation at 37 °C, the ΔpakA conidia remained viable (unpublished data).
To identify other factors involved in the pakA-dependent differences in germination at 25 °C and 37 °C, conidial germination was analyzed in detail at both temperatures in strains carrying mutations in cflA (CDC42 orthologue), gasC (GPA2 orthologue), and rasA (RAS2 orthologue), which have been shown previously to affect germination at 25 °C but which had not been characterized at 37 °C [22–24]. The role of CflA, GasC, and RasA in germination at 37 °C was characterized by assessing the percentage of germinated conidia after 8, 12, 15, and 20 h at both 25 °C and 37 °C in two strains of each genotype (Tables S1 and S2). Two-level nested ANOVA was performed on the data for each time point at both 25 °C and 37 °C to test if germination rates differed significantly between genotypes and also between transformants of the same genotype (Materials and Methods). ANOVA showed there was a significant difference between genotypes in all time points except 0 h. In a few instances, there was variation within transformants of the same genotype (Tables S1 and S2). Strains expressing the dominant negative cflAD120A allele showed delayed germination at both 25 °C and 37 °C (Figure 6A and 6C). Dominant activated cflAG14V strains displayed accelerated germination at both 25 °C and 37 °C (Figure 6B and 6D). These results indicate that active CflA promotes conidial germination at both 25 °C and 37 °C. Likewise, the dominant interfering gasCG207R strains showed delayed germination at both 25 °C and 37 °C (Figure 7A and 7C). The dominant activated gasCG45R strains showed accelerated germination at 25 °C and 37 °C, suggesting that, like CflA, GasC is required for conidial germination at both 25 °C and 37 °C (Figure 7B and 7D). Both the dominant activated (rasAG19V) and dominant negative (rasAD125A) rasA strains showed a decrease in germination at 25 °C (Table S1). In contrast, both the dominant negative and dominant activated strains showed wild type germination patterns at 37 °C, suggesting that RasA is required for conidial germination at 25 °C but not at 37 °C (Table S2).
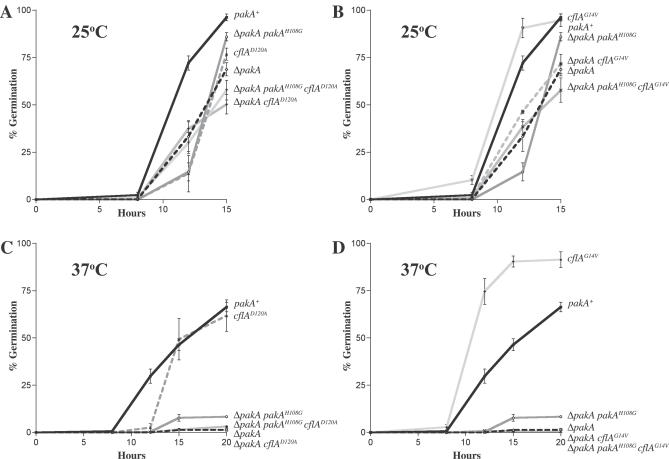
Figure 6. PakA Is Required for CflA-Dependent Germination at Both 25 °C and 37 °C.
The percentage of germinated conidia was measured in wild-type (pakA+), cflAD120A, ΔpakA, ΔpakA pakAH108G, ΔpakA cflAD120A, and ΔpakA pakAH108G cflAD120A strains after incubation at 25 °C for 8, 12, and 15 h (A) and at 37 °C for 8, 12, 15, and 20 h (C). The percentage of germinated conidia was measured in wild-type (pakA+), cflAG14V, ΔpakA, ΔpakA pakAH108G, ΔpakA cflAG14V, and ΔpakA pakAH108G cflAG14V strains after incubation at 25 °C for 8, 12, and 15 h (B) and at 37 °C for 8, 12, 15, and 20 h (D). One representative strain of each genotype is shown. Three independent experiments were performed and standard error of the mean bars are shown. (A, B) At 25 °C, the ΔpakA and ΔpakA pakAH108G strains show a delay in germination when compared with the wild-type control. (A) Compared with the wild type (pakA+), the cflAD120A strain shows delayed germination at 25 °C. The ΔpakA cflAD120A and ΔpakA pakAH108G cflAD120A strains show delayed germination similar to the ΔpakA and ΔpakA pakAH108G strains. (B) Compared with wild type (pakA+), the cflAG14V strain shows accelerated germination. The ΔpakA cflAG14V and ΔpakA pakAH108G cflAG14V strains show delayed germination similar to the ΔpakA and ΔpakA pakAH108G strains. (C, D) At 37 °C, the ΔpakA and ΔpakA pakAH108G strains display a dramatic reduction in the number of germinated conidia when compared with wild type (pakA+). (C) Compared with the wild type (pakA+), the cflAD120A strain shows delayed germination at 37 °C. The ΔpakA cflAD120A and ΔpakA pakAH108G cflAD120A strains show delayed germination similar to the ΔpakA strain. (D) In contrast to wild type (pakA+), the cflAG14V strain exhibits accelerated germination at 37 °C. The ΔpakA cflAG14V and ΔpakA pakAH108G cflAG14V strains show dramatically reduced germination at 37 °C similar to the ΔpakA and pakAH108G strains.
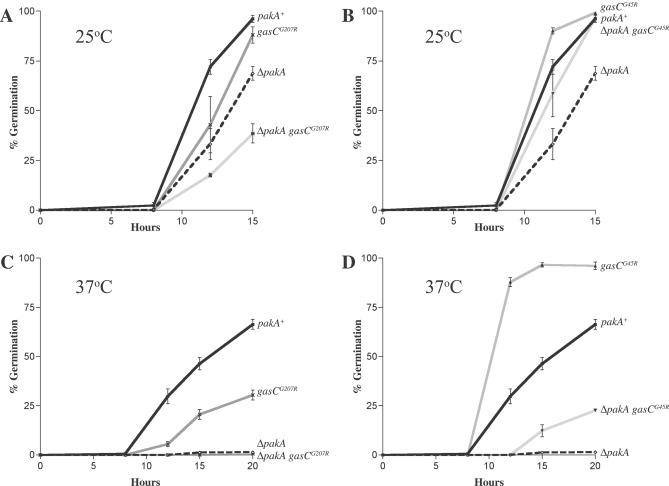
Figure 7. Expression of the Dominant Activated gasCG45R Allele Partially Suppresses the Germination Defect of the ΔpakA Mutant.
The percentage of germinated conidia was measured in the wild-type (pakA+), ΔpakA, gasCG207R, gasCG45R, ΔpakA gasCG207R, ΔpakA gasCG45R strains after incubation at 25 °C for 8, 12, and 15 h (A and B) and at 37 °C for 8, 12, 15, and 20 h (C, D). One representative strain of each genotype is shown. Three independent experiments were performed. Standard error of the mean bars are shown. Compared with wild type (pakA+), the ΔpakA mutant has delayed germination at 25 °C (A, B). (A) At 25 °C, the gasCG207R strains show delayed germination compared with wild type. ΔpakA gasCG207R strains display delayed germination that is lower than the ΔpakA and gasCG207R single mutant strains. (B) At 25 °C, the gasCG45R strains show accelerated germination compared with wild type. The ΔpakA gasCG45R strains exhibit germination kinetics similar to wild type. (C, D) The ΔpakA mutant has a severe germination defect at 37 °C. (C) The gasCG207R strains show delayed germination at 37 °C. ΔpakA gasCG207R strains display severely reduced germination similar to the ΔpakA strain. (D) Compared with wild type (pakA+), gasCG45R strains show accelerated germination at 37 °C. ΔpakA gasCG45R strains show germination that is intermediate between wild type and the ΔpakA strain.
Genetic Interactions Between cflA, pakA, and rasA during Conidial Germination
To investigate any genetic interaction between pakA and cflA, double mutants were generated (ΔpakA cflA+, ΔpakA cflAD120A, ΔpakA cflAG14V, ΔpakA pakAH108G cflA+, ΔpakA pakAH108G cflAD120A, and ΔpakA pakAH108G cflAG14V) and germination was characterized by assessing the percentage of germinated conidia after 8, 12, 15, and 20 h at both 25 °C and 37 °C (Tables S1 and S2). It should be noted that multiple copy integrants may result in significant overexpression. Two strains of each genotype were assessed and a single representative strain is shown in Figure 6. ANOVA was performed on the data for each time point at both 25 °C and 37 °C to test if germination rates differed significantly between genotypes and also between transformants of the same genotype (Materials and Methods). ANOVA showed there was a significant difference between genotypes at all time points except 0 h. In a few instances, there was variation within transformants of the same genotype (Tables S1 and S2). The control ΔpakA cflA+ and ΔpakA pakAH108G cflA+ strains showed germination patterns at 25 °C and 37 °C that were indistinguishable from the parental ΔpakA and ΔpakA pakAH108G strains. At 25 °C, the ΔpakA cflAD120A and ΔpakA pakAH108G cflAD120A strains displayed delayed conidial germination at rates similar to the single cflAD120A mutant strains (Figure 6A). At 37 °C, the ΔpakA cflAD120A and ΔpakA pakAH108G cflAD120A strains displayed dramatically reduced germination like the single ΔpakA and ΔpakA pakAH108G strains (Figure 6C).
In contrast to the accelerated germination observed in cflAG14V mutants at 25 °C, which is much faster than wild-type (Figure 6B), the ΔpakA cflAG14V and ΔpakA pakAH108G cflAG14V double mutants display slower than wild-type germination at 25 °C, similar to the ΔpakA and ΔpakA pakAH108G single mutant strains (Figure 6B). This suggests that the accelerated germination observed in cflAG14V strains at 25 °C requires active PakA and an interaction of CflA and PakA via the PakA CRIB domain.
Likewise at 37 °C, in contrast to the accelerated germination observed in cflAG14V mutants, the ΔpakA cflAG14V and ΔpakA pakAH108G cflAG14V double mutants display dramatically reduced germination at rates equivalent to the ΔpakA and ΔpakA pakAH108G single mutant strains (Figure 6D). The inability of the cflAG14V dominant activated allele to suppress the reduced germination phenotype of the ΔpakA and ΔpakA pakAH108G mutants suggests that at 37 °C, CflA acts upstream of PakA during germination and that an interaction between CflA and the CRIB domain of PakA is required for germination to proceed.
P. marneffei RasA operates upstream of CflA at both 25 °C and 37 °C [23]. The genetic interaction of pakA and rasA was investigated by generating ΔpakA rasA+, ΔpakA rasAD125A, and ΔpakA rasAG19V double mutants. At 25 °C, the ΔpakA rasA+ strains showed wild-type germination patterns, whereas the ΔpakA rasAD125A and ΔpakA rasAG19V mutants showed delayed germination similar to the single rasAD125A and rasAG19V mutants (Table S1). At 37 °C, the ΔpakA rasA+, ΔpakA rasAD125A, and ΔpakA rasAG19V double mutants showed severely reduced germination similar to the ΔpakA mutant (Table S2).
GasC Activates Two Pathways Regulating Conidial Germination at 25 °C and 37 °C
To investigate the genetic interaction between pakA and gasC, double mutants were generated (ΔpakA gasC+, ΔpakA gasCG207R, and ΔpakA gasCG45R). Germination was characterized in five strains of each double mutant genotype by assessing the percentage of germinated conidia after 8, 12, 15, and 20 h at both 25 °C and 37 °C (Tables S1 and S2). ANOVA was performed on the data for each time point at both 25 °C and 37 °C (Materials and Methods) and revealed there was a significant difference between genotypes at all time points except 0 h. In a few instances, there was variation within transformants of the same genotype (Tables S1 and S2). ΔpakA gasC+ strains were indistinguishable from ΔpakA. At 25 °C, ΔpakA gasCG207R strains have delayed germination, which is slower than both the gasCG207R single mutant strains and the ΔpakA mutant (Figure 7A). At 37 °C ΔpakA gasCG207R strains have a severe reduction in germination similar to ΔpakA (Figure 7C). In contrast to the accelerated germination of the gasCG45R single mutant strains and the delayed germination of the ΔpakA mutant, the ΔpakA gasCG45R double mutant strains show wild-type germination at 25 °C (Figure 7B). In addition, ΔpakA gasCG45R double mutant strains show germination rates at 37 °C, which are lower than those of wild-type and the gasCG45R mutants but higher than that of the ΔpakA mutant (Figure 7D). This indicates that expression of the gasCG45R mutant allele partially suppresses the germination defects of ΔpakA and suggests that GasC regulates two pathways during germination, one of which is independent of PakA.
Discussion
Conserved Proteins Initiate Filamentous Growth but the Downstream Effectors Are Differentially Regulated
The establishment of an axis of polarised growth is orchestrated by the asymmetric distribution of cellular components through the localisation and activation of proteins required for growth. Some of the core components regulating polarised growth establishment in S. cerevisiae are conserved both in the genome and functionally during polarity establishment in more complex organisms. However, the question remains as to how multi-cellular organisms generate the greater diversity of distinct cell types with the same set of core components. Unlike small eukaryotes like fungi, larger eukaryotes such as flies and mammals often have an increased number of factors involved in polarity establishment with significant redundancy [26–28]. One possible mechanism is to alter the activity of the key establishment proteins in different cell types, while another is to differentially regulate the effector proteins. In the dimorphic pathogen P. marneffei, the germination of conidia gives rise to two different developmental pathways and cell types. The regulation of conidial germination by CflA (CDC42 orthologue) and GasC (GPA2 orthologue) at both 25 °C and 37 °C suggests that the core components regulating polarised growth establishment in S. cerevisiae may be conserved during polarity establishment in germinating conidia of filamentous fungi. However, mutations in pakA, a potential downstream effector of cflA, result in a dramatic reduction in the rates of germination at 37 °C but not 25 °C. This suggests that in P. marneffei conserved polarity establishment proteins regulate germination, but the downstream effectors are differentially regulated to give rise to distinct cell types. The results suggest a model in which, at 25 °C, GasC activates two pathways regulating conidial germination (Figure 8). In one pathway, RasA activates CflA, which activates PakA—by association via the CRIB domain—and a proposed additional effector to establish polarised hyphal growth (Figure 8). At 37 °C, GasC also activates two pathways regulating conidial germination. In one pathway, similar to 25 °C, CflA activates PakA via the CRIB domain, and this interaction is required for PakA function. Unlike 25 °C, RasA does not activate CflA, PakA plays a crucial role during the establishment of polarised arthroconidiating hyphal growth, and no additional effector is required (Figure 8).
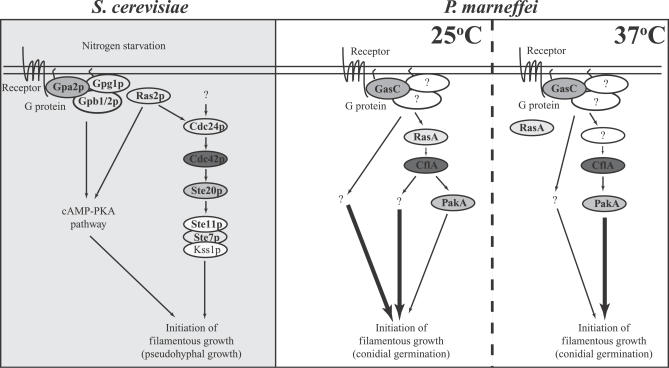
Figure 8. Models for the Regulation of the Initiation of Filamentous Growth in S. cerevisiae and P. marneffei .
(A) In S. cerevisiae, nitrogen starvation triggers a switch from the unicellular budding growth form to a filamentous growth form (pseudohyphal growth). The activated GTPase Ras2p, activates the GEF Cdc24p, which catalyses the activation of the Rho GTPase Cdc42p. Cdc42p activates the PAK kinase Ste20p, which leads to the activation of a MAPK kinase cascade, which culminates in the activation of the transcription factor Ste12p. The activation of a heterotrimeric G protein is also involved in the initiation of filamentous growth in S. cerevisiae, via activation of cAMP signaling.
(B) In P. marneffei, GasC (Gpa2p homologue) activates two pathways regulating conidial germination at 25 °C. In one pathway, GasC activates RasA (Ras2p homologue), which activates CflA (Cdc42p homologue). CflA activates PakA (Ste20p homologue) via the CRIB domain and this interaction is required for PakA function. CflA also activates an additional unknown effector at 25 °C, which plays a more important role than PakA in germination. It is possible that GasC directly acts on PakA and does not act upstream of RasA and CflA. B. At 37 °C, GasC also activates two pathways regulating conidial germination. In one pathway, GasC activates CflA, which activates PakA. Similar to 25 °C, at 37 °C CflA activates PakA via the CRIB domain and this interaction is required for PakA function. Unlike 25 °C, at 37 °C RasA is not required for germination, PakA plays a crucial role during the establishment of polarised arthroconidiating hyphal growth, and no additional effector is required.
The G-protein GasC Regulates Two Signaling Pathways Required for the Initiation of Filamentous Growth in P. marneffei
gasC encodes a heterotrimeric guanine nucleotide-binding (G-protein) α-subunit with homology to S. cerevisiae GPA2. Gpa2p is required for the initiation of filamentous growth (pseudohyphal growth) in response to nitrogen starvation via the activation of the cAMP-PKA pathway [2,3]. In P. marneffei, expression of dominant activated gasCG45R and dominant interfering gasCG207R alleles at both 25 °C and 37 °C results in accelerated or delayed conidial germination, respectively, and suggests that GasC acts as a general upstream activator initiating filamentous growth. This study suggests that GasC activates two pathways regulating germination at both 25 °C and 37 °C, one of which is dependent on PakA (Figure 8). In contrast to the accelerated germination of gasCG45R strains and the slightly delayed germination of ΔpakA, ΔpakA gasCG45R double mutant strains display wild-type germination at 25 °C. A reduction in the germination rates suggests that PakA is acting downstream of GasC at 25 °C. However, as germination of the ΔpakA gasCG45R strains is higher than ΔpakA, GasC must also regulate an additional PakA-independent pathway activating germination at 25 °C. This hypothesis is supported by the germination rates observed in the ΔpakA gasCG207R strains, which are lower than both the single mutant strains, suggesting these mutations have an additive effect. Likewise, at 37 °C, partial suppression of the ΔpakA germination defect by expression of the gasCG45R allele indicates that GasC acts as an upstream regulator of two pathways activating conidial germination, one of which is dependent on PakA. The activation of two pathways regulating the initiation of filamentous growth in P. marneffei differs from S. cerevisiae. In S. cerevisiae, the Gpa2p regulation of pseudohyphal growth occurs via the activation of the cAMP-PKA pathway, which is independent of the Ste20p/MAPK cascade [3]. The pseudohyphal growth defect of a gpa2 mutant can be suppressed by addition of cAMP or overexpression of the dominant activated RAS2G19V allele. However, the pseudohyphal defect cannot be suppressed by the overexpression of the dominant active STE11–4 allele and has no effect on expression of a reporter gene (FRE-lacZ), which is known to be regulated by the MAP kinase cascade [3]. These results together suggest that, unlike P. marneffei GasC, S. cerevisiae, Gpa2p regulates the cAMP pathway but not the Ste20p-MAPK cascade, thus regulating the initiation of filamentous growth.
The Cdc42 Orthologue in P. marneffei Activates PakA during the Initiation of Filamentous Growth
In S. cerevisiae Cdc42p localises and activates Ste20p (reviewed in [12]). Therefore P. marneffei PakA is a potential downstream effector of the CflA (Cdc42p orthologue) and may be involved in similar processes. Like cflAD120A strains, which show delayed germination at 37 °C, the ΔpakA mutant exhibits a dramatic reduction in conidial germination at 37 °C, suggesting that PakA is crucial during the establishment of polarised growth to give rise to arthroconidiating hyphae. However, unlike CflA, PakA plays only a minor role during germination at 25 °C, as strains expressing the dominant negative cflAD120A allele show a severe delay in germination compared with only a slight delay in conidial germination for the ΔpakA and ΔpakA pakAH108G strains. These results suggest that PakA acts downstream of CflA at both 25 °C and 37 °C in P. marneffei. This hypothesis is also supported by the observation that the accelerated germination seen in cflAG14V strains is abrogated by the ΔpakA at both 25 °C and 37 °C in ΔpakA cflAG14V strains.
PAKs contain a conserved N-terminal CRIB domain and a C-terminal kinase domain (reviewed in [27]). The CRIB domain of S. cerevisiae Ste20p negatively inhibits the kinase domain preventing signaling [25]. This autoinhibition is relieved by interaction of the CRIB domain with Cdc42p, and this interaction is also necessary for localisation of Ste20p to sites of polarised growth [25]. The H345G mutation in the Ste20p CRIB domain shows reduced interaction with Cdc42p in a two-hybrid assay and the loss of correct localisation to sites of polarised growth. The phenotype of the STE20H345G strain is almost equivalent to the null, indicating that the interaction of Cdc42p and Ste20p is essential for Ste20p function (and for relief of autoinhibition of the kinase domain) [25]. The H108G CRIB domain mutation in P. marneffei is equivalent to the H345G of S. cerevisiae and was found to have a similar effect. The H108G mutation resulted in reduced localisation of PakA to sites of polarised growth and a phenotype equivalent to the deletion mutant. Compared with the accelerated germination of cflAG14V single mutant strains at both 25 °C and 37 °C, the ΔpakA pakAH108G cflAG14V mutant strains exhibited reduced germination, suggesting that the interaction of CflA and PakA via the CRIB domain is required during conidial germination at both 25 °C and 37 °C. The interaction of CflA and PakA is also required during polarised growth of yeast cells, as the ΔpakA pakAH108G strains showed a swollen, abnormal yeast morphology at 37 °C, similar to the deletion strain. In S. cerevisiae, the CRIB domain is essential for pseudohyphal growth but dispensable for G protein–mediated pheromone signaling [11]. In P. marneffei the CRIB domain of PakA is also required for the initiation of filamentous growth, but unlike S. cerevisiae the initiation of filamentous growth in P. marneffei by PakA is partially dependent on G-protein signaling.
The minor role played by PakA during germination and hyphal growth at 25 °C suggests that another CflA effector, possibly a Cla4p orthologue, is required for these processes. The genomes of A. nidulans, M. grisea, U. maydis, Coprinopsis cinerea, Neurospora crassa, and C. albicans encode two PAKs, one with homology to Ste20p and the other to Cla4p (http://www.broad.mit.edu/annotation/fgi/). In addition to the CRIB and kinase domains of Ste20p orthologues, Cla4p homologues have a pleckstrin homology domain. Cla4p in S. cerevisiae is required for septation but does not play a role in pseudohyphal growth. However, deletion of CLA4 in the yeasts Yarrowia lipolytica and C. albicans blocks filament formation, and the cla4 deletion mutant of the plant pathogen U. maydis is unable to form filaments during infection [29–32].
PAKs in P. marneffei May Have Overlapping Roles during Polarised Growth of Hyphae at 25 °C
cflA has been previously shown to play a pivotal role during hyphal morphogenesis with mutations resulting in grossly aberrant hyphae [24]. It was therefore expected that the ΔpakA strain may have a similar phenotype, albeit less severe, as CflA is proposed to interact and activate numerous effector proteins. The lack of a ΔpakA hyphal phenotype suggests that PakA is not required for hyphal growth. Like P. marneffei, deletion of the Ste20p homologues in C. albicans and U. maydis does not result in defects in hyphal morphology [33,34]. However, the co-localisation of the GFP::PakA fusion protein with actin and the localisation to the same cellular locations as CflA at nascent septation sites and to the hyphal apex suggests a role during polarised growth of hyphae [23]. In S. cerevisiae, Ste20p directly phosphorylates Bni1p, a component of the polarisome [35]. The polarisome is a protein complex, which contains Bni1p, Spa2p, Pea2p, and Bud6p, that promotes polarised morphogenesis during filamentous growth [35,36]. The Bni1p homologue, SepA, plays a conserved role in polarised growth in A. nidulans [37]. However, A. nidulans lacks a Pea2p homologue and the Spa2 homologue, SpaA, is only partially conserved in sequence and function, indicating that the polarisome in filamentous fungi likely consists of a modified set of components with different contributions to polarisome function [36]. The implication is that in P. marneffei, in addition to specific developmental roles, pakA and pakB play complementary, and possibly overlapping, roles in the establishment of polarised growth during conidial germination and in the maintenance of an axis of polarisation during hyphal growth. How the two PAKs coordinately regulate different aspects of development of multi-cellular fungi still remains unclear. The analysis of a CLA4 orthologue from P. marneffei may resolve many of these issues.
Materials and Methods
Molecular techniques.
P. marneffei genomic DNA and RNA was isolated as previously described [38,39]. Southern and northern blotting was performed with Amersham Hybond N+ membrane according to the manufacturer's instructions. Filters were hybridized using [α−32P]dATP-labeled probes by standard methods [40].
Cloning and plasmid construction.
Primers L18 (5′-TGATCCCACAAAACTTTACT-3′) and L19 (5′-GCTCGTTTCTCAGGGTCCAC-3′) were used to amplify the A. nidulans genomic sequence encoding the conserved kinase domain of the STE20 homologue. The PCR product was sequenced and used to screen a P. marneffei genomic library (constructed in λGEM-11) at low stringency (50% formamide, 2 x SSC, 37 °C). A 6.4 kb NotI/BglII hybridizing fragment from a positively hybridizing clone was subcloned into NotI/BamHI digested pBluescript II SK+ (pKB5751). Sequencing was performed by the Australian Genome Research Facility and analyzed using Sequencher 3.1.1 (Gene Codes Corporation). The Genbank accession number of the P. marneffei pakA gene is AY621630.
A pakA deletion construct (pKB5792) was generated by replacing the 2.5 kb EcoRV/ClaI fragment of pKB5751 with the 2.5 kb SmaI/ClaI fragment containing the pyrG+ selectable marker. This resulted in pyrG+ flanked by 2.6 kb of 5′ and 1.1 kb of 3′ pakA sequence, and deleted from −425 to +2030. Inverse PCR using the mutagenic primers N30 (5′-ACATGAGTAACACCGACAGGG-3′) and N32 (5′-TGGATACGACAATCAGACTGG-3′) was used to introduce the H108G mutation into pakA generating pKB5908. The integrity of the construct was confirmed by sequencing.
The gfp::pakA and gfp::pakAH108G constructs were generated by ligating a BamHI/XbaI fragment from pKB5751 (pakA) and pKB5908 (pakAH108G) into pALX196 (gpdA(p)::gfp).
Fungal strains and media.
Strains used in this study are shown in Table 1. The ΔpakA strain (ΔpakA::pyrG+) was generated by transforming the strain SPM4 with linearised pKB5792 and selecting for pyrG+. Transformation was performed using the previously described protoplast method [38]. The ΔpakA pyrG− strain was isolated by plating the ΔpakA strain (ΔpakA::pyrG+) on medium containing 1 mg/mL−1 5-FOA supplemented with 10 mM γ-amino butyric acid (GABA) and 5 mM uracil to select for the loss of the pyrG marker. A 5-FOA resistant sector was isolated that had a restriction pattern consistent with loss of pyrG at the pakA locus. The strain is unable to grow in the absence of 5 mM uracil.
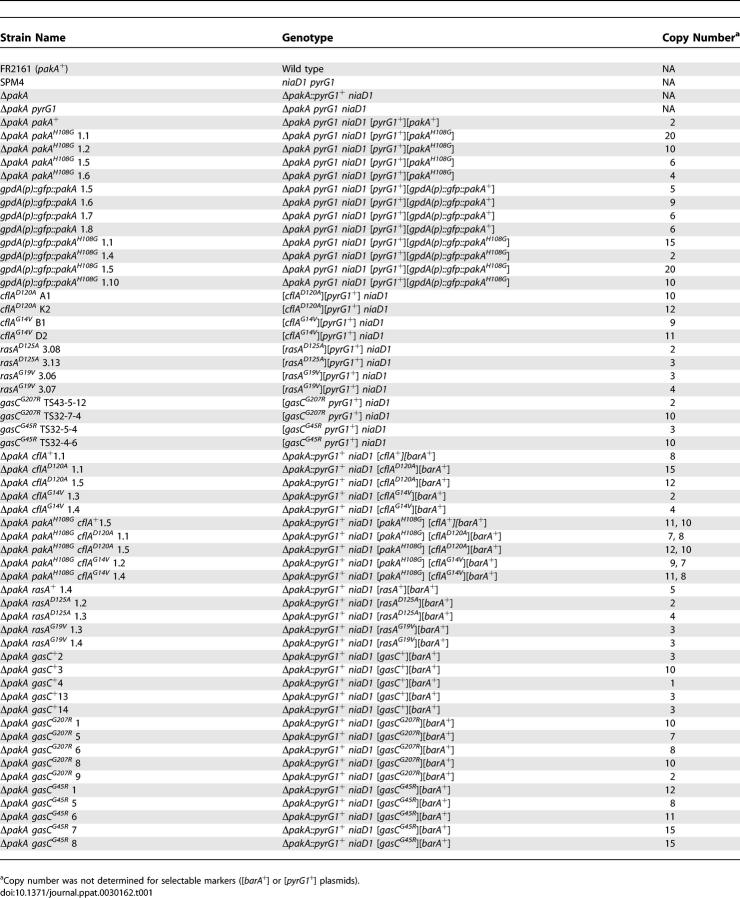
Table 1.
P. marneffei Strains Used in This Study
P. marneffei FRR2161, SPM4, cflAD120A, cflAG14V, rasAD125A, rasAG19V, gasCG207R, and gasCG45R have been previously described [22–24,38]. All other strains listed in Table 1 were generated by cotransformation of the ΔpakA pyrG or ΔpakA strain with plasmids containing the appropriate mutant allele and either pAB4342 (pyrG+) or pMT1612 (barA+) as selectable markers. Southern blot analysis was used to confirm cotransformation and to determine the plasmid copy number.
At 25 °C strains were grown on A. nidulans minimal medium (ANM) supplemented with 1% glucose and 10 mM GABA or on synthetic dextrose (SD) medium supplemented with 10 mM ammonium sulphate ([NH4]2SO4) as a sole nitrogen source [41,42]. At 37 °C, strains were grown on BHI medium or on SD medium supplemented with 10 mM (NH4)2SO4.
In vivo macrophage assay.
J774 murine macrophages (1 × 105) were seeded into each well of a 6-well microtitre tray containing one sterile coverslip and 2 mL of complete Dulbecco's Modified Eagle Medium (complete DMEM: DMEM, 10% fetal bovine serum, 2 mM L-glutamine and penicillin-streptomycin). Macrophages were incubated at 37 °C for 24 h before activation with 0.1μg/mL−1 lipopolysaccharide (LPS) from E. coli (Sigma). Macrophages were incubated a further 24 h at 37 °C and washed 3 times in phosphate buffered saline, and 2 mL of complete DMEM medium containing 1 × 106 conidia was added. A control lacking conidia was also performed. Macrophages were incubated for 2 h at 37 °C (to allow conidia to be engulfed), washed once in phosphate buffered saline (to remove free conidia), and incubated a further day at 37 °C. Macrophages were fixed in 4% paraformaldehyde and stained with 1 mg/mL−1 fluorescent brightener 28 (calcofluor, CAL) to observe fungal cell walls. The numbers of germinated conidia was measured microscopically by counting the numbers of germinated conidia (conidia with a visible germ tube or yeast cells) in a population of approximately 100. Three independent experiments were performed. Mean and standard error of the mean values were calculated using GraphPad Prism3.
Microscopy.
P. marneffei strains were grown on slides covered with a thin layer of solid medium, with one end resting in liquid medium [38]. Wild-type (pakA+), ΔpakA, ΔpakA pakA+, and ΔpakA pakAH108G strains were grown on ANM medium supplemented with GABA at 25 °C for 2 or 4 d. At 37 °C, strains were grown on BHI medium for 4 d or in liquid BHI medium for 6 d.
Immunofluorescence microscopy for examination of the actin cytoskeleton was performed using standard protocols [43]. Double staining was performed with the mouse C4 monoclonal anti-actin (Chemicon International) and rabbit anti-GFP polyclonal primary antibodies, as well as ALEXA 488 rabbit anti-mouse (Molecular Probes) and ALEXA 594 anti-rabbit (Molecular Probes) secondary antibodies. Single immunostaining controls and a minus primary antibody control were also performed. The gfp::pakA and gfp::pakAH108G strains were grown on agar-coated slides containing ANM plus GABA for 2 d or SD with 5 mM ammonium tartrate (NH4T) for 4 d at 37 °C.
Slides were examined using differential interference contrast (DIC) and epifluorescence optics for GFP, antibody fluorescence, cell wall staining with fluorescent brightener 28 (calcofluor, CAL), or nuclear staining with DAPI and viewed on a Reichart Jung Polyvar II microscope. Images were captured using a SPOT CCD camera (Diagnostic Instruments) and processed in Adobe Photoshop.
Germination experiments.
Approximately 106 spores were inoculated into 300 μL of SD plus 10 mM (NH4)2SO4 and incubated for 8, 12, or 15 h at 25 °C or for 8, 12, 15, or 20 h at 37 °C. The rates of germination were measured microscopically by counting the numbers of germinating conidia (conidia with a visible germ tube) in a population of 100. Three independent experiments were performed. Mean and standard error of the mean values were calculated using GraphPad Prism3. Two-level nested ANOVA was performed on the data for each time point at both 25 °C and 37 °C to test if germination rates differed significantly between genotypes and between transformants of the same genotype. ANOVA simultaneously tests two null hypotheses; there is no difference between the means of the data sets from all genotypes and there is no difference between the means of the data sets between transformants of the same genotype. The generation of two F-statistics and probability values allow rejection or acceptance of these null hypothesis at a 99% confidence. Values with an asterisk in Tables S1 and S2 showed significant differences between transformants of the same genotype.
To investigate the differential regulation of conidial germination at 25 °C and 37 °C, wild-type and ΔpakA conidia were incubated in liquid medium at different temperatures ranging from 25 °C to 37 °C. The 20-h incubation was performed in a gradient thermocycler using 2 °C temperature increments from 25 °C to 37 °C. The media was then transferred to a microtitre tray and the rates of germination were measured microscopically by counting the numbers of germinating conidia (conidia with a visible germ tube) in a population of 100. Three independent experiments were performed. Mean and standard error of the mean values were calculated using GraphPad Prism3.
Supporting Information
(41 KB DOC)
(54 KB DOC)
Abbreviations
- 5-FOA
5-fluoroorotic acid
- ANM
A. nidulans minimal medium
- BHI
brain heart infusion
- CAL
calcofluor
- cAMP
cyclic adenosine monophosphate
- CRIB
Cdc42/Rac interactive binding
- DAPI
4′6-diamidino-2-phenylindole
- DIC
differential interference contrast
- DMEM
Dulbecco's Modified Eagle Medium
- GEF
guanine nucleotide exchange factor
- LPS
lipopolysaccharide
- MAPK
mitogen-activated protein kinase
- PAK
p21-activated kinase
- PKA
protein kinase A
- SD
synthetic dextrose
Footnotes
Author contributions. KJB and AA conceived and designed the experiments, analyzed the data, contributed reagents/materials/analysis tools, and wrote the paper. KJB performed the experiments.
Funding. This work was supported by grants to A. Andrianopoulos from the Australian Research Council, the National Health and Medical Research Council and the Howard Hughes Medical Institute. A. Andrianopoulos is a Howard Hughes Medical Institute International Scholar.
Competing interests. The authors have declared that no competing interests exist.
References
- Mosch HU, Roberts RL, Fink GR. Ras2 signals via the Cdc42/Ste20/mitogen-activated protein kinase module to induce filamentous growth in Saccharomyces cerevisiae . Proc Natl Acad Sci U S A. 1996;93:5352–5356. doi: 10.1073/pnas.93.11.5352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubler E, Mosch HU, Rupp S, Lisanti MP. Gpa2p, a G-protein alpha-subunit, regulates growth and pseudohyphal development in Saccharomyces cerevisiae via a cAMP-dependent mechanism. J Biol Chem. 1997;272:20321–20323. doi: 10.1074/jbc.272.33.20321. [DOI] [PubMed] [Google Scholar]
- Lorenz MC, Pan X, Harashima T, Cardenas ME, Xue Y, et al. The G protein–coupled receptor Gpr1 is a nutrient sensor that regulates pseudohyphal differentiation in Saccharomyces cerevisiae . Genetics. 2000;154:609–622. doi: 10.1093/genetics/154.2.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harashima T, Heitman J. The Galpha protein Gpa2 controls yeast differentiation by interacting with kelch repeat proteins that mimic Gbeta subunits. Mol Cell. 2002;10:163–173. doi: 10.1016/s1097-2765(02)00569-5. [DOI] [PubMed] [Google Scholar]
- Harashima T, Heitman J. Galpha subunit Gpa2 recruits kelch repeat subunits that inhibit receptor–G protein coupling during cAMP-induced dimorphic transitions in Saccharomyces cerevisiae . Mol Biol Cell. 2005;16:4557–4571. doi: 10.1091/mbc.E05-05-0403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harashima T, Anderson S, Yates JR, III, Heitman J. The kelch proteins Gpb1 and Gpb2 inhibit Ras activity via association with the yeast RasGAP neurofibromin homologs Ira1 and Ira2. Mol Cell. 2006;22:819–830. doi: 10.1016/j.molcel.2006.05.011. [DOI] [PubMed] [Google Scholar]
- Peeters T, Louwet W, Geladé R, Nauwelaers D, Thevelein JM, Versele M. Kelch-repeat proteins interacting with the Galpha protein Gpa2 bypass adenylate cyclase for direct regulation of protein kinase A in yeast. Proc Natl Acad Sci U S A. 2006;103:13034–13039. doi: 10.1073/pnas.0509644103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versele M, de Winde JH, Thevelein JM. A novel regulator of G protein signalling in yeast, Rgs2, downregulates glucose-activation of the cAMP pathway through direct inhibition of Gpa2. EMBO J. 1999;18:5577–5591. doi: 10.1093/emboj/18.20.5577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo S, Ma P, Cauwenberg L, Winderickx J, Crauwels M, Teunissen A, et al. Involvement of distinct G-proteins, Gpa2 and Ras, in glucose- and intracellular acidification–induced cAMP signalling in the yeast Saccharomyces cerevisiae . EMBO J. 1998;17:3326–3341. doi: 10.1093/emboj/17.12.3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Cerione R, Bender A. Control of the yeast bud-site assembly GTPase Cdc42. Catalysis of guanine nucleotide exchange by Cdc24 and stimulation of GTPase activity by Bem3. J Biol Chem. 1994;269:2369–2372. [PubMed] [Google Scholar]
- Leberer E, Wu C, Leeuw T, Fourest-Lieuvin A, Segall JE, et al. Functional characterization of the Cdc42p binding domain of yeast Ste20p protein kinase. EMBO J. 1997;16:83–97. doi: 10.1093/emboj/16.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DI. Cdc42: an essential Rho-type GTPase controlling eukaryotic cell polarity. Microbiol Mol Biol Rev. 1999;63:54–105. doi: 10.1128/mmbr.63.1.54-105.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Whiteway M, Thomas DY, Leberer E. Molecular characterization of Ste20p, a potential mitogen-activated protein or extracellular signal-regulated kinase kinase (MEK) kinase kinase from Saccharomyces cerevisiae . J Biol Chem. 1995;270:15984–15992. doi: 10.1074/jbc.270.27.15984. [DOI] [PubMed] [Google Scholar]
- Cook JG, Bardwell L, Thorner J. Inhibitory and activating functions for MAPK Kss1 in the S. cerevisiae filamentous-growth signalling pathway. Nature. 1997;390:85–88. doi: 10.1038/36355. [DOI] [PubMed] [Google Scholar]
- Drogen F, O'Rourke SM, Stucke VM, Jaquenoud M, Neiman AM, et al. Phosphorylation of the MEKK Ste11p by the PAK-like kinase Ste20p is required for MAP kinase signaling in vivo. Curr Biol. 2000;10:630–639. doi: 10.1016/s0960-9822(00)00511-x. [DOI] [PubMed] [Google Scholar]
- Harris SD. Morphogenesis is coordinated with nuclear division in germinating Aspergillus nidulans conidiospores. Microbiology. 1999;145:2747–2756. doi: 10.1099/00221287-145-10-2747. [DOI] [PubMed] [Google Scholar]
- Momany M, Westfall PJ, Abramowsky G. Aspergillus nidulans swo mutants show defects in polarity establishment, polarity maintenance and hyphal morphogenesis. Genetics. 1999;151:557–567. doi: 10.1093/genetics/151.2.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osherov N, May G. Conidial germination in Aspergillus nidulans requires RAS signaling and protein synthesis. Genetics. 2000;155:647–656. doi: 10.1093/genetics/155.2.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- d'Enfert C, Fontaine T. Molecular characterization of the Aspergillus nidulans treA gene encoding an acid trehalase required for growth on trehalose. Mol Microbiol. 1997;24:203–216. doi: 10.1046/j.1365-2958.1997.3131693.x. [DOI] [PubMed] [Google Scholar]
- Fillinger S, Chaveroche MK, Shimizu K, Keller N, d'Enfert C. cAMP and ras signalling independently control spore germination in the filamentous fungus Aspergillus nidulans . Mol Microbiol. 2002;44:1001–1016. doi: 10.1046/j.1365-2958.2002.02933.x. [DOI] [PubMed] [Google Scholar]
- Andrianopoulos A. Control of morphogenesis in the human fungal pathogen Penicillium marneffei . Int J Med Microbiol. 2002;292:331–347. doi: 10.1078/1438-4221-00217. [DOI] [PubMed] [Google Scholar]
- Zuber S, Hynes MJ, Andrianopoulos A. The G-protein alpha-subunit GasC plays a major role in germination in the dimorphic fungus Penicillium marneffei . Genetics. 2003;164:487–499. doi: 10.1093/genetics/164.2.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyce KJ, Hynes MJ, Andrianopoulos A. The Ras and Rho GTPases genetically interact to co-ordinately regulate cell polarity during development in Penicillium marneffei . Mol Microbiol. 2005;55:1487–1501. doi: 10.1111/j.1365-2958.2005.04485.x. [DOI] [PubMed] [Google Scholar]
- Boyce KJ, Hynes MJ, Andrianopoulos A. The CDC42 homolog of the dimorphic fungus Penicillium marneffei is required for correct cell polarization during growth but not development. J Bact. 2001;183:3447–3457. doi: 10.1128/JB.183.11.3447-3457.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamson RE, Winters MJ, Pryciak PM. Cdc42 regulation of kinase activity and signaling by the yeast p21-activated kinase Ste20. Mol Cell Biol. 2002;22:2939–2951. doi: 10.1128/MCB.22.9.2939-2951.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sells MA, Chernoff J. Emerging from the Pak: the p21-activated protein kinase family. Trends Cell Biol. 1997;7:162–167. doi: 10.1016/S0962-8924(97)01003-9. [DOI] [PubMed] [Google Scholar]
- Dan I, Watanabe NM, Kusumi A. The Ste20 group kinases as regulators of MAP kinase cascades. Trends Cell Biol. 2001;11:220–230. doi: 10.1016/s0962-8924(01)01980-8. [DOI] [PubMed] [Google Scholar]
- Jaffe AB, Hall A. Rho GTPases: biochemistry and biology. Annu Rev Cell Dev Biol. 2005;21:247–269. doi: 10.1146/annurev.cellbio.21.020604.150721. [DOI] [PubMed] [Google Scholar]
- Kohler JR, Fink GR. Candida albicans strains heterozygous and homozygous for mutations in mitogen-activated protein kinase signaling components have defects in hyphal development. Proc Natl Acad Sci U S A. 1996;93:13223–13228. doi: 10.1073/pnas.93.23.13223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leberer E, Ziegelbauer K, Schmidt A, Harcus D, Dignard D, et al. Virulence and hyphal formation of Candida albicans require the Ste20p-like protein kinase CaCla4p. Curr Biol. 1997;7:539–546. doi: 10.1016/s0960-9822(06)00252-1. [DOI] [PubMed] [Google Scholar]
- Leveleki L, Mahlert M, Sandrock B, Bolker M. The PAK family kinase Cla4 is required for budding and morphogenesis in Ustilago maydis . Mol Microbiol. 2004;54:396–406. doi: 10.1111/j.1365-2958.2004.04296.x. [DOI] [PubMed] [Google Scholar]
- Szabo R. Cla4 protein kinase is essential for filament formation and invasive growth of Yarrowia lipolytica . Mol Genet Genomics. 2001;265:172–179. doi: 10.1007/s004380000405. [DOI] [PubMed] [Google Scholar]
- Leberer E, Harcus D, Broadbent ID, Clark KL, Dignard D, et al. Signal transduction through homologs of the Ste20p and Ste7p protein kinases can trigger hyphal formation in the pathogenic fungus Candida albicans . Proc Natl Acad Sci U S A. 1996;93:13217–13222. doi: 10.1073/pnas.93.23.13217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DG, Garcia-Pedrajas MD, Hong W, Yu Z, Gold SE, et al. An ste20 homologue in Ustilago maydis plays a role in mating and pathogenicity. Eukaryot Cell. 2004;3:180–189. doi: 10.1128/EC.3.1.180-189.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goehring AS, Mitchell DA, Tong AH, Keniry ME, Boone C, et al. Synthetic lethal analysis implicates Ste20p, a p21-activated potein kinase, in polarisome activation. Mol Biol Cell. 2003;14:1501–1516. doi: 10.1091/mbc.E02-06-0348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virag A, Harris SD. Functional characterization of Aspergillus nidulans homologues of Saccharomyces cerevisiae Spa2 and Bud6. Eukaryot Cell. 2006;5:881–895. doi: 10.1128/EC.00036-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpless KE, Harris SD. Functional characterization and localization of the Aspergillus nidulans formin SepA. Mol Biol Cell. 2002;13:469–479. doi: 10.1091/mbc.01-07-0356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borneman AR, Hynes MJ, Andrianopoulos A. The abaA homologue of Penicillium marneffei participates in two developmental programmes: conidiation and dimorphic growth. Mol Microbiol. 2000;38:1034–1047. doi: 10.1046/j.1365-2958.2000.02202.x. [DOI] [PubMed] [Google Scholar]
- Boyce KJ, Hynes MJ, Andrianopoulos A. Control of morphogenesis and actin localization by the Penicillium marneffei RAC homolog. J Cell Sci. 2003;116:1249–1260. doi: 10.1242/jcs.00319. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning, a laboratory manual. Cold Spring Harbor (New York): Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore RE, Seidman JA, et al. Current protocols in molecular biology. New York: John Wiley and Sons, Inc; 1994. [Google Scholar]
- Cove DJ, Quatrano RS, Hartmann E. The alignment of the axis of asymmetry in regenerating protoplasts of the moss, Ceratodon purpureus, is determined independently of axis polarity. Development. 1996;122:371–379. doi: 10.1242/dev.122.1.371. [DOI] [PubMed] [Google Scholar]
- Fischer R, Timberlake WE. Aspergillus nidulans apsA encodes a coiled-coil protein required for nuclear positioning and completion of asexual development. J Cell Biol. 1995;128:485–498. doi: 10.1083/jcb.128.4.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(41 KB DOC)
(54 KB DOC)