Abstract
Tomato (Solanum lycopersicum) Pto encodes a protein kinase that confers resistance to bacterial speck disease. A second protein kinase, Pti1, physically interacts with Pto and is involved in Pto-mediated defense signaling. Pti1-related sequences are highly conserved among diverse plant species, including rice (Oryza sativa), but their functions are largely unknown. Here, we report the identification of a null mutant for the Pti1 homolog in rice and the functional characterization of Os Pti1a. The rice pti1a mutant was characterized by spontaneous necrotic lesions on leaves, which was accompanied by a series of defense responses and resistance against a compatible race of Magnaporthe grisea. Overexpression of Pti1a in rice reduced resistance against an incompatible race of the fungus recognized by a resistance (R) protein, Pish. Plants overexpressing Pti1a were also more susceptible to a compatible race of the bacterial pathogen Xanthomonas oryzae pv oryzae. These results suggest that Os Pti1a negatively regulates defense signaling for both R gene–mediated and basal resistance. We also demonstrated that repression of the rice RAR1 gene suppressed defense responses induced in the pti1a mutant, indicating that Pti1a negatively regulates RAR1-dependent defense responses. Expression of a tomato Pti1 cDNA in the rice pti1a mutant suppressed the mutant phenotypes. This contrasts strikingly with the previous finding that Sl Pti1 enhances Pto-mediated hypersensitive response (HR) induction when expressed in tobacco (Nicotiana tabacum), suggesting that the molecular switch controlling HR downstream of pathogen recognition has evolved differently in rice and tomato.
INTRODUCTION
Plants have evolved surveillance and defense response systems to protect themselves from pathogen attack. The first step of defense against attempted microbial invasion is achieved by a pattern recognition receptor that detects a pathogen-associated molecular pattern (PAMP) (Zipfel and Felix, 2005; Chisholm et al., 2006). Pathogenic microbes have specialized systems that suppress or evade plant PAMP-triggered defenses and facilitate tissue invasion by secreting several effector proteins (Nurnberger et al., 2004; Chisholm et al., 2006). When a plant resistance (R) protein directly or indirectly recognizes a specific pathogen effector, which is often the product of a pathogen avirulence (avr) gene, the plant exhibits heightened defense against the pathogen (Jones and Takemoto, 2004; Jones and Dangl, 2006). The recognition of different pathogens by several R proteins appears to amplify a common set of defense responses and triggers rapid and strongly localized generation of reactive oxygen species, pathogen-related (PR) gene expression, and accumulation of antimicrobial compounds (Dangl and Jones, 2001; Durrant and Dong, 2004). These responses are often accompanied by localized programmed cell death known as the hypersensitive response (HR) at the site of pathogen invasion (Greenberg and Yao, 2004).
In the last decade, a large number of R genes from several species have been identified by map-based cloning, insertional mutagenesis, or various high-throughput methods (Hammond-Kosack and Parker, 2003). Sequence comparisons among these genes reveal a remarkable conservation of structural features, despite the diversity of the pathogens which their products recognize (Nimchuk et al., 2003). The largest class of R genes, termed the NB-LRR class, encodes a cytoplasmic protein with a Leu-rich repeat (LRR) and a nucleotide binding (NB) site. Although the signal components downstream of R proteins are thought to be conserved, only a few components that regulate the fundamental aspects of R protein–triggered responses have been isolated (Hammond-Kosack and Parker, 2003). Among those identified, RAR1 (required for Mla12 resistance), HSP90 (heat shock protein 90), and SGT1 (suppressor of the G2 allele of skp1) are required for resistance mediated by various NB-LRR R proteins (Shirasu and Schulze-Lefert, 2003; Piffanelli et al., 2004). The RAR1 protein is required by particular R proteins that are effective against bacterial, oomycete, and viral pathogens reported in barley (Hordeum vulgare), Arabidopsis thaliana, and tobacco (Nicotiana tabacum) (Freialdenhoven et al., 1994; Liu et al., 2002; Muskett et al., 2002; Tornero et al., 2002). It interacts with both HSP90 and SGT1 and is considered to function as a molecular chaperon, in association with HSP90, to stabilize certain R proteins (Azevedo et al., 2002; Takahashi et al., 2003).
To date, there have been several reports suggesting a link between PAMP-triggered basal resistance and R protein–mediated resistance at the molecular level. In Arabidopsis, RIN4 is a negative regulator of PAMP signaling and is targeted by Pseudomonas syringae type-III effector AvrRpt2 for degradation, leading to the activation of an R protein, RPS2 (Kim et al., 2005). AvrB, a P. syringae effector protein, suppresses PAMP-triggered immunity through RAR1, which is indispensable for the stabilization of RPM1, the R protein corresponding to AvrB (Shang et al., 2006). rar1 mutations in Arabidopsis allowed enhanced growth of the virulent bacterial strain P. syringae DC3000 (Holt et al., 2005). Nb SGT1 is required not only for R protein–mediated HR induction but also for some non-host resistance responses (Peart et al., 2002). These observations suggest that the signaling pathways for PAMP-triggered immunity and R protein–mediated race-specific resistance substantially share common regulatory components.
The tomato (Solanum lycopersicum) R protein Pto confers race-specific resistance to the bacterial pathogen P. syringae pv tomato carrying the avirulence effector proteins AvrPto or AvrPtoB (Pedley and Martin, 2003). The Pto gene encodes a Ser/Thr protein kinase and is unique among several classes of known R proteins. Pto was shown to directly interact with both the bacterial effector proteins in a yeast two-hybrid system assay. However, little is known about the signal transduction mechanism downstream of the recognition event. A number of potential downstream components of the Pto signaling pathway have been reported, such as the protein kinase Pti1, and transcription factors Pti4, Pti5, and Pti6 (Pedley and Martin, 2003). Pti1 interacts with Pto and is phosphorylated by Pto in vitro. Overexpression of Pti1 in tobacco causes enhanced HR in leaves when challenged with P. syringage pv tabaci expressing AvrPto (Zhou et al., 1995). However, there is no direct evidence to support the involvement of Pti1 in Pto-mediated disease resistance.
Despite considerable efforts to find and characterize gain-of-function or loss-of-function mutants in several plant species, it is still unclear how R proteins transmit signals to downstream factors, what the limiting factors in evoking defense responses might be, or how the relationship between basal resistance and race-specific resistance is established. To develop more insight into plant defense signaling, we screened for mutants that had enhanced resistance to rice (Oryza sativa) blast disease from a collection of mutant lines generated by rice endogenous retrotransposon Tos17 insertion (Hirochika et al., 2004). Here, we describe a rice mutant that develops spontaneous lesions on its leaves and has enhanced resistance. This increased resistance was caused by depletion of Os Pti1a, a tomato Pti1 homolog. Overexpression of Os Pti1a in transgenic rice plants impaired resistance in both compatible and incompatible interactions. Moreover, we demonstrated that the mutant phenotype, including acquired resistance, lesion formation, and PR gene expression, was suppressed by silencing the expression of a rice ortholog of RAR1 (Os RAR1). We propose that Os Pti1a plays a role in the negative regulation of both R protein–mediated defense signaling and basal resistance.
RESULTS
Identification of the ttm1 Mutant and Its Phenotype
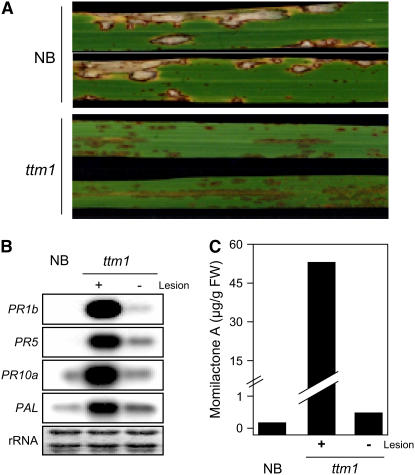
We identified a lesion mimic mutant (ND5001) among stable insertion mutant lines of Japonica rice cv Nipponbare (NB) produced by endogenous retrotransposon Tos17 (Hirochika, 2001; Hirochika et al., 2004) and designated it ttm1 (for Tos17 triggered mutation1). The ttm1 lesion is recessively inherited, and homozygous ttm1 plants have stunted growth with spontaneous small and obscure lesions over both leaf surfaces (Figures 1A and 1B). Lesions appeared at ∼30 d after sowing in the field and after ∼40 to 50 d after sowing in a greenhouse. The variation in the timing of lesion appearance may be due to differences in growth conditions. Because the lesion pattern was similar to that seen during HR, we presumed that lesion formation results from induction of the HR pathway triggered by the ttm1 mutation. To examine whether the ttm1 mutation activates defense responses, we inoculated mutant and wild-type plants with the rice blast fungus Magnaporthe grisea. The cultivar NB has Pish, an R protein that is active against rice blast fungus isolates containing avrPish, such as race 102.0 (incompatible), but not against race 003.0 (compatible) (Imbe and Matsumoto, 1985). Disease resistance of the ttm1 mutant line against the incompatible race was comparable to that of NB (data not shown). However, ttm1 plants exhibited strong resistance against the compatible race after lesion formation (Figure 2A) but not before lesion formation (data not shown). The ttm1 plants expressed defense-related genes, PR1b, PR5, PR10a, and PAL, after the appearance of lesions (Figure 2B). Furthermore, momilactone A, the major phytoalexin of rice (Cartwright et al., 1977), accumulated in uninfected leaves with lesions to a level ∼150-fold higher than NB at the same developmental stage but was negligible in ttm1 leaves without lesions (Figure 2C). Thus, a series of defense responses were activated in the ttm1 mutant at developmental stages after the appearance of lesions. These results suggest that the causative gene of the ttm1 mutant negatively regulates the defense signaling leading to HR induction.
Figure 1.
Phenotype of the ttm1 Mutant.
(A) ttm1 and NB plants were grown in the field and photographed at the early flowering stage. These plants were transplanted to pots for photography.
(B) Lesion phenotype of ttm1 and NB on young leaves at the early flowering stage.
Figure 2.
Defense-Related Phenotypes of ttm1.
(A) NB and ttm1 were inoculated with a compatible race (003.0) of M. grisea. Lesions are shown on leaf blades 10 d after inoculation.
(B) Total RNA was extracted from lesion-negative (−) and lesion-positive (+) leaves of ttm1 mutants and NB. RNA gel blots of 10 μg total RNA were hybridized with radiolabeled probes of defense-related genes as indicated. rRNA is shown by staining with methylene blue as a loading control.
(C) A rice phytoalexin, momilactone A, was extracted from lesion-negative (−) and lesion-positive (+) leaves of ttm1 mutants and NB and quantified by the method described in Methods. FW, fresh weight.
Cloning of Ttm1
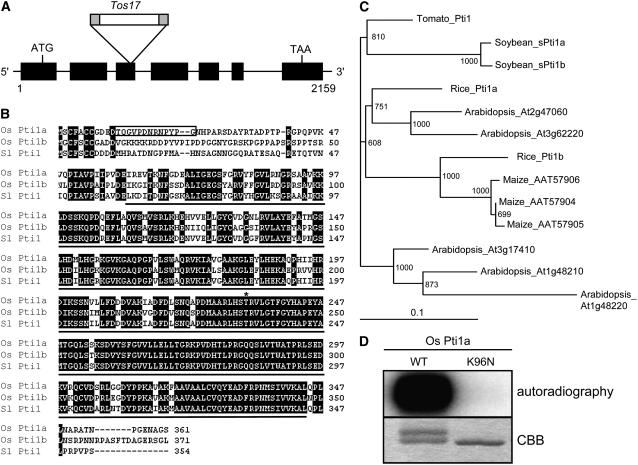
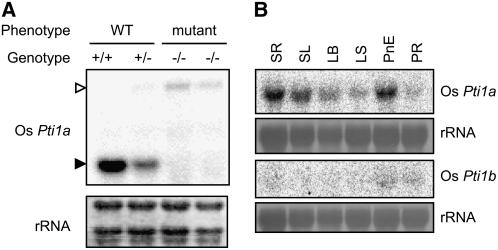
To isolate the Ttm1 allele, we extracted genomic DNA from the progeny of ttm1 heterozygotes and subjected it to DNA gel blot analysis to examine the cosegregation of Tos17 with the mutant phenotype. Genomic DNA flanking cosegregating Tos17 was isolated by thermal asymmetric interlaced PCR with a Tos17-specific primer and degenerate primers (Liu et al., 1995). The PCR product was used as a probe for DNA gel blot hybridization to confirm the cosegregation with the lesion mimic phenotype. As expected, only the plants with the mutant phenotype carried a homozygous insertion (data not shown). A search of the GenBank nucleotide database using the BLAST program with the flanking sequence revealed that Tos17 was inserted in the third exon of Os Pti1a (Figure 3A). This gene had high similarity to Sl Pti1 (87% similarity at the amino acid level). Sl Pti1 encodes a cytoplasmic protein kinase and was originally identified by a yeast two-hybrid screen as a protein that interacts with Pto, which is a tomato R protein to P. syringae pv tomato, the causative agent of bacterial speck disease (Zhou et al., 1995). Because tomato Pti1 is phosphorylated by Pto, but Pto is not phosphorylated by Pti1, Sl Pti1 seems to function downstream of Pto in a phosphorylation cascade (Sessa et al., 2000). The predicted Os Pti1a gene would encode 361 amino acids, and its deduced Mr was 39.3 kD. RNA gel blots showed that Os Pti1a transcripts were not detected in Os pti1a (ttm1) homozygous mutants but were detected in wild-type and Os pti1a heterozygous mutants (Figure 4A). Therefore, Os pti1a is a null mutation. A database search also revealed that there is another homolog of Sl Pti1 in rice. It was designated Os Pti1b, and its predicted product has an 83% similarity to Sl Pti1 and 81% to Os Pti1a (Figures 3B and 3C). An amino acid sequence alignment of deduced Pti1 proteins showed that the protein kinase domain was highly conserved, but the N-terminal regions were highly variable (Figure 3B). The Thr residue at 233 (Thr-233) in Sl Pti1, which is the major site phosphorylated by Pto, was conserved in both Os Pti1a and Os Pti1b (Sessa et al., 2000). The highly similar protein sequences of Os Pti1a and Os Pti1b suggest that these proteins are functionally redundant. Nevertheless, the disruption of just one of them, Os Pti1a, was adequate to trigger spontaneous cell death and defense responses. To further explore this observation, we analyzed the transcript levels of Os Pti1a and Os Pti1b (Figure 4B). Pti1a was detected abundantly in the roots, young leaves, adult leaves, and preemergent panicles but not in ripening panicles. By contrast, Pti1b transcripts were barely detectable in each of the organs tested. The negligible expression of Pti1b is thus the likely reason that the loss of Pti1a is sufficient to trigger cell death and defense responses despite the presence of paralog Pti1b. To examine Pti1b function, we isolated an Os pti1b knockout Tos17 insertion line (ND4512). This line grew as healthily as the wild type without lesion formation and did not exhibit any enhanced resistance (data not shown). We then crossed this line with the Os pti1a knockout line to produce a pti1a pti1b double knockout mutant. The double mutant was morphologically and developmentally indistinguishable from the pti1a single mutant (data not shown). These results suggest that Os Pti1b has only a minor function, if any, which is consistent with its extremely low levels of expression.
Figure 3.
Comparison of Os Pti1a and Related Proteins.
(A) Relative position of the Tos17 insertion within the Os Pti1a gene. Exons are indicated by the black boxes.
(B) Alignment of the predicted amino acid sequences of Os Pti1a, Os Pti1b, and Sl Pti1. A black line below Sl Pti1 indicates a conserved protein kinase domain, and the box delimits the peptide sequence used to derive Os Pti1a–specific antisera. The asterisk marks the T233 phosphorylation site of Sl Pti1 by Pto and the corresponding sites in Os Pti1a and Pti1b.
(C) A phylogenetic tree constructed with the amino acid sequences of kinase domain of Os Pti1a and Pti1 family members from several plant species.
(D) Autophosphorylation assay for Os Pti1a (WT) and its mutant form (K96N) in vitro. The upper band in the wild-type lane seen in the Coomassie blue (CBB)–stained gel is presumably the phosphorylated form.
Figure 4.
Transcript Analysis of Pti1a and Pti1b in Rice.
(A) Total RNA was extracted from homozygous and heterozygous pti1a plants and NB. Ten micrograms of total RNA was used for RNA gel blot analysis. The closed arrowhead indicates normal size of Os Pti1a mRNA, and the open arrowhead indicates the size of Os Pti1a with Tos17. Equal loading of RNA samples is shown by the quantity of rRNA.
(B) Expression analysis of Os Pti1a and Os Pti1b was performed with total RNA extracted from seedling root (SR), seedling leaf (SL), adult leaf blade (LB), adult leaf sheath (LS), panicle, not emerged (PnE), and panicle, ripening (PR).
Os Pti1a Is a Functional Protein Kinase
To determine whether the protein encoded by Os Pti1a is a functional protein kinase, it was expressed as a polyhistidine-tagged protein in Escherichia coli. Incubation of the purified fusion protein with [γ-32P]ATP in an in vitro kinase assay showed that Os Pti1a was capable of strong autophosphorylation (Figure 3D). The K96N mutation, which is known to completely abolish the autophosphorylation activity of Sl Pti1 (Zhou et al., 1995), abolished the autophosphorylation activity of Os Pti1a (Figure 3D), indicating that Os Pti1a encodes a functional protein kinase similar to Sl Pti1.
Os Pti1a and Sl Pti1 Complement the Os pti1a Mutant Phenotype
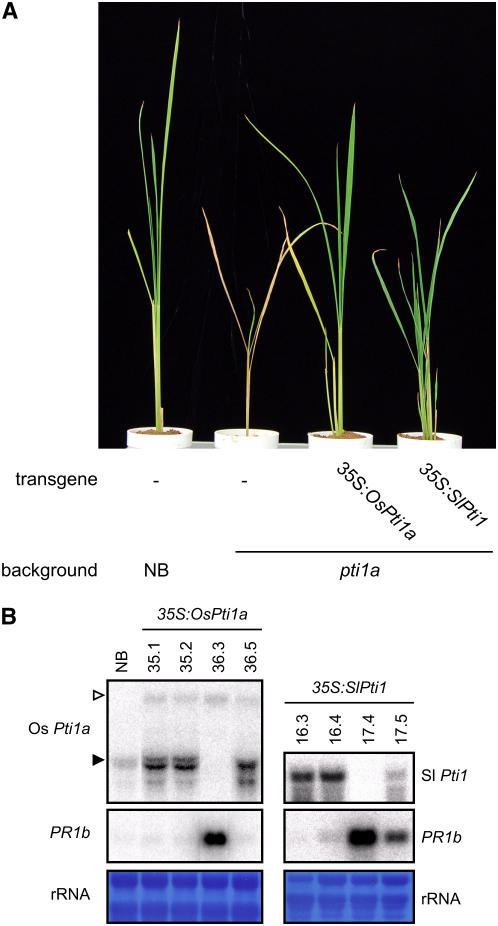
To confirm that the null mutation of Os Pti1a causes lesion formation and the induction of defense reactions, we screened for other allelic mutants from Tos17 insertion mutant lines by a PCR-based method using specific primers for Tos17 and Os Pti1a. Unfortunately, however, no allelic mutants were identified. We then transformed a full-length cDNA of Os Pti1a under the control of the cauliflower mosaic virus 35S promoter into the pti1a homozygous mutant. We obtained three independent transgenic lines and used their T1 and T2 generations for the following analysis. None of the transgenic plants was stunted, nor did they have lesions, and they were as healthy as the wild type (Figure 5A). The upregulation of PR1b expression observed in the pti1a line was lost when Pti1a was overexpressed in this mutant (Figure 5B). These transgenic lines were not resistant to a compatible race of M. grisea, unlike the background pti1a mutant, indicating that the expression of Os Pti1a cDNA complemented the pti1a mutant phenotypes. The loss of Pti1a function thus results in Os pti1a mutant phenotypes.
Figure 5.
Complementation of the pti1a Mutation with Os Pti1a and Sl Pti1 cDNA.
(A) cDNA of the Os Pti1a or Sl Pti1 genes under control of the 35S promoter was introduced into the Os pti1a homozygous mutant. The photograph was taken 40 d after sowing of transgenic plants (T1 generation) and control plants.
(B) Total RNA was extracted from NB and the pti1a homozygous mutant with the introduced constructs 35S:Os Pti1a or 35S:Sl Pti1 (T1 generation). Ten micrograms of total RNA was hybridized with radiolabeled probes as indicated. The closed arrowhead indicates normal size of Os Pti1a mRNA, and the open arrowhead indicates the size of Os Pti1a with Tos17. Equal loading of RNA samples is shown by the quantity of rRNA.
Transgenic tobacco plants that overexpress Sl Pti1 cDNA show enhanced HR in leaves when challenged with P. syringae pv tabaci strains carrying the avirulence gene avrPto, suggesting that Sl Pti1 functions as a positive regulator of Pto-mediated cell death and disease resistance (Zhou et al., 1995). This contrasts strikingly with the observed phenotypes of the Os pti1a mutant. To investigate this apparent discrepancy, we expressed Sl Pti1 cDNA in the Os pti1a homozygous mutant under the control of the 35S promoter. Interestingly, the expression of Sl Pti1 cDNA blocked lesion formation and PR1b expression in the Os pti1a mutant (Figures 5A and 5B). Similar results were obtained with other three independent lines. These results indicate that the two Pti1 proteins, Sl Pti1 and Os Pti1a, are functionally equivalent. Nevertheless, the contrasting phenotypes of the mutant and transgenic plants indicate that a downstream molecular switch controlling HR has evolved differently in monocotyledonous rice and dicotyledonous tomato.
Os Pti1a Overexpression Reduces Plant Resistance
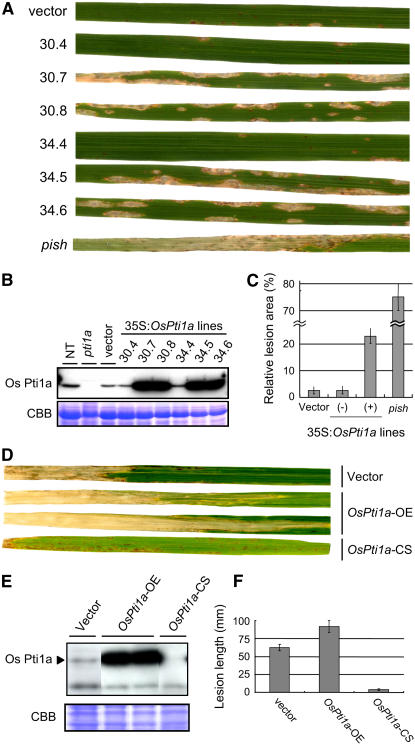
As described, the Os pti1a mutant produces spontaneous lesions resulting from the activation of defense responses in the absence of any pathogenic signal. This result implies that Os Pti1a is involved in the negative regulation of the defense signaling pathway. This prompted us to overexpress full-length Os Pti1a cDNA (Os Pti1a-OE) under the control of the 35S promoter in the NB background to determine what effect it would have on defense responses with rice blast fungus. Accumulation of Os Pti1a proteins in the T1 generation was measured using a specific antibody against the nonconserved N-terminal region of Pti1a (Figure 6B). Pti1a protein was detected in non-transgenic and vector control (25.3) plants but not in the pti1a homozygous mutant, confirming that this antibody is specific to Os Pti1a. We selected three individuals each from the T1 progeny of two independent transgenic lines (30 and 34) and inoculated them with an incompatible M. grisea race carrying avrPish (Figure 6A). The vector control plants developed small HR lesions on the leaves. The lesions seen on leaves of transgenic plants with high Pti1a protein levels (30.7, 30.8, 34.5, and 34.6) were significantly larger than those on their siblings with lower levels of Pti1a accumulation (30.4 and 34.4) and a vector control. To evaluate disease severity in these lines, we compared total lesion area per leaf (Figure 6C). As a negative control, we used the pish null mutant, which was derived from Tos17-induced NB mutant lines (our unpublished data). Lesion areas on the Os Pti1a-OE lines were significantly larger than those on the vector control, but they were still restricted compared with the pish mutant. Thus, the overaccumulation of Os Pti1a proteins partially impaired the disease resistance triggered by Pish. Similar results were obtained with other three independent lines. To examine how general the regulatory role of Os Pti1a is in incompatible rice blast interactions, we tested another gene-for-gene system in NB mediated by Pi19 that is effective against M. grisea carrying avr19 (Hayashi et al., 1998). However, we did not observe any reduction in defense reactions in Os Pti1a-OE lines against this incompatible race (see Supplemental Figure 1 online). Pi19-mediated defense response is much stronger than that mediated by Pish; therefore, it is likely that the effects of Os Pti1a overexpression were masked by the effective defense reaction. However, we cannot exclude the possibility that Os Pti1a is not involved in all R protein–mediated defense responses.
Figure 6.
Effect of Overexpression of Os Pti1a on Disease Resistance.
(A) Three-week-old plants (T1 generation) derived from transgenic NB lines carrying 35S:Os Pti1a (30.3 to 34.6: derived from two independent transgenic lines, 30 and 34) or empty vector (vector) and pish mutant were challenged with an incompatible race (102.0) of rice blast fungus by spray inoculation. In two siblings (30.4 and 34.4), the transgene 35S:Os Pti1a was segregated out. The picture is of leaf blades 7 d after inoculation.
(B) Total protein was extracted from T1 plants derived from transgenic NB lines carrying 35S:Os Pti1a or empty vector (vector) and the pti1a mutant. Os Pti1a protein was detected with anti-Os Pti1a antibody raised against the peptide shown in Figure 3B. NT indicates untransformed NB control plants.
(C) Quantitative lesion area data for leaves of NB with 35S:Os Pti1a or vector and pish mutant. The plus sign indicates transgenic plants in which Os Pti1a was overaccumulated. The minus sign indicates siblings with a lower level of Os Pti1a accumulation. Results from three independent experiments are presented as average ± sd (n = 7 to 10).
(D) Representative leaves showing the extent of lesion development 21 d after inoculation with the bacterial pathogen X. oryzae pv oryzae (race 1) in leaves of generation T2 overexpression lines (Os Pti1a-OE), cosuppression lines (Os Pti1a-CS), and vector control.
(E) Os Pti1a protein in total protein was detected with anti-Os Pti1a antibody. The bottom panel shows the Coomassie blue–stained gel.
(F) Lesion length measurements for the T2 generation of each line. The graphs depict the mean ± sd from three independent experiments (n > 20).
Effects of Os Pti1a Overexpression on Compatible Pathogen Interactions
To investigate the effects of Os Pti1a overexpression on compatible interactions, we first tested a compatible race of the rice blast fungus (race 003.0). However, there was no enhanced susceptibility in Os Pti1a-OE lines to this race of the fungus (data not shown). Presumably, the effect of Os Pti1a overexpression on the compatible interaction is too small to be detectable against the strong pathogenicity of this fungus. We then tested the rice pathogen Xanthomonas oryzae pv oryzae (Xoo), the causal agent of rice bacterial blight disease. Because wild-type NB exhibits moderate levels of resistance against compatible races of Xoo (race 1), this pathogen seemed to provide a suitable system to assess the effect of Os Pti1a overexpression on compatible pathogen interactions. We used the T2 generation of Os Pti1a-OE plants for the inoculation of Xoo, after examining Pti1a protein levels by immunoblotting (Figure 6E). Xoo-induced lesions were 1.5-fold longer in the leaves of Os Pti1a-OE lines than in the vector control plants (Figures 6D and 6F), indicating that Pti1a suppresses basal resistance against the bacteria. The Os pti1a null mutant has severe growth defects, which made it difficult to evaluate its resistance against Xoo. Among 35S:Os Pti1a lines, however, we were able to obtain Os Pti1a-cosuppressed lines (Os Pti1a-CS) that did not exhibit growth defects. In Os Pti1a-CS lines, Pti1a protein accumulated at the levels comparable with those in Os Pti1a-OE lines during early developmental stages (data not shown). The proteins declined to negligible levels as the plants developed (Figure 6E), suggesting that the cosuppression occurred in the middle of development. This delayed occurrence of cosupression is a likely mechanism for the normal growth of the cosuppressed lines. When tested after Os Pti1a protein levels had declined, Os Pti1a-CS plants showed strong resistance against Xoo compared with wild-type and Os Pti1a-OE lines. These results indicate that Os Pti1a also has a negative effect on rice plant resistance to compatible pathogens.
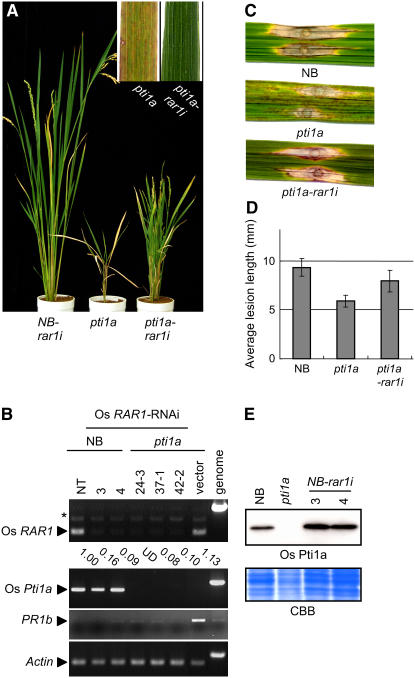
Os Pti1a Negatively Regulates RAR1-Mediated Defense Signaling
The recognition of pathogen invasion by R proteins is followed by rapid activation of the defense signal cascade. Some regulatory proteins involved in gene-for-gene resistance have been characterized in Arabidopsis, barley, and tobacco. One such regulatory protein, RAR1, is required for the functioning of various R proteins and acts upstream of HR induction (Shirasu et al., 1999). To examine the genetic interaction between Pti1a and RAR1 in rice, we silenced Os RAR1 expression by RNA interference (RNAi) in the Os pti1a homozygous mutant (Os pti1a-rar1i) and in NB (NB-rar1i). RT-PCR analysis demonstrated that RAR1 transcript levels decreased to ∼10% of the wild type in both Os pti1a-rar1i and NB-rar1i (Figure 7B). Silencing of RAR1 in NB caused no visible phenotype without pathogen challenge; however, the effect of RAR1 silencing was striking in Os pti1a-rar1i plants. In these plants, the spontaneous lesion formation due to pti1a mutation was completely abolished. In addition, RT-PCR revealed that the expression of PR1b was reduced in pti1a-rar1i lines compared with the pti1a control line carrying empty vector (Figure 7B). The dwarf phenotype of the pti1a mutant was also suppressed, although incompletely. Similar phenotypes were observed in >10 pti1a-rar1i plants from four independent lines. These results indicate that rar1i is genetically epistatic to Os pti1a.
Figure 7.
RAR1 is Required for pti1a-Induced Defense Responses in Rice.
(A) Representative phenotypes of 3-month-old transgenic rice (T1 generation) carrying the Os RAR1-RNAi interference construct or ospti1a homozygous mutant.
(B) RT-PCR analysis was performed on total RNA isolated from leaves of T1 progeny of transgenic rice carrying Os RAR1-RNAi or empty vector. NT indicates untransformed control NB plants. RT-PCR of an actin gene was used as a control for RNA template amounts. The RNA level of Os RAR1 was quantified by image analysis and is shown relative to actin. UD, undetectable.
(C) Five-week-old NB, pti1a, or pti1a-rar1i was inoculated with a compatible race (003.0) of M. grisea. The photograph was taken 10 d after inoculation.
(D) Average lesion length of the fungus disease. The graphs depict the mean ± sd obtained from four to six measurements using at least each two independent transgenic lines.
(E) Total protein was extracted from NB, pti1a, and NB-rar1i transformants. Os Pti1a protein was detected with anti-Os Pti1a antibody. The bottom panel shows the Coomassie blue–stained gel.
To investigate RAR1 dependence of cell death induction in other lesion mimic mutants, we suppressed Os RAR1 in rice mutants cdr1 and cdr2 (Takahashi et al., 1999), which are characterized by spontaneous cell death and a series of defense responses similar to the pti1a mutant. Os RAR1 suppression, however, affected neither the spatial pattern nor the timing of lesion formation in cdr mutants (see Supplemental Figure 2 online), indicating that RAR1-dependent cell death induction is not general with lesion mimic mutants.
In rice, RAR1 silencing did not compromise the resistance mediated by three R genes for blast fungus (N.P. Thao, L. Chen, A. Nakashima, S. Hara, K. Umemura, A. Takahashi, K. Shirasu, T. Kawasaki, and K. Shimamoto, unpublished data). These observations are in agreement with previous reports that RAR1 is not required for the functioning of all R genes (Shirasu and Schulze-Lefert, 2003). We found that both NB-rar1i and Os pti1a-rar1i plants retained resistance against incompatible blast fungus races carrying avrPish (data not shown), indicating that Os RAR1 is not required for Pish-mediated resistance either. Then, we examined the effects of RAR1 silencing on the enhanced resistance of the pti1a mutant against a compatible race of blast fungus. The enhanced resistance against the compatible race of blast fungus observed in the pti1a mutant was largely cancelled in Os pti1a-rar1i plants (Figures 7C and 7D). These results clearly indicate that the function of Os Pti1a in the negative regulation of blast resistance is also dependent on Os RAR1.
Recent biochemical studies suggesting various requirements for RAR1 in an R protein–triggered signaling pathway seem to reflect its role as a protein chaperone to stabilize or protect R protein complexes from degradation (Nimchuk et al., 2003; Jones and Takemoto, 2004). If Os Pti1a functions in close proximity to R proteins, the stability of the Os Pti1a protein could be regulated by a chaperone activity involving RAR1. To examine this hypothesis, we measured the levels of Pti1a protein accumulation in NB-rar1i. However, we found no significant difference in Pti1a levels between NB and NB-rar1i plants (Figure 7D), indicating that the stability of Os Pti1a protein does not depend on RAR1 activity.
DISCUSSION
Sl Pti1 was originally identified as a protein that interacts with an R protein Pto in tomato. Because Sl Pti1 is phosphorylated by Pto and encodes a Ser/Thr protein kinase, it is thought that Sl Pti1 functions downstream of Pto and transmits a defense signal to downstream components through its protein kinase activity (Pedley and Martin, 2003). However, the genetic data supporting a direct involvement of Sl Pti1 in Pto-dependent disease resistance is very limited. Here, we provide genetic evidence that a rice homolog of Pti1, Os Pti1a, negatively regulates both R protein–mediated resistance and basal resistances in a Os RAR1–dependent manner.
Os Pti1a Functions as a Negative Regulator of R Protein–Mediated and Basal Resistance
Loss of Os Pti1a induced resistance against compatible races of both M. grisea and Xoo (Figures 2 and 6D). Overexpression of Os Pti1a reduced Pish-mediated resistance to an incompatible race of the fungus. Os Pti1a overexpression also reduced basal resistance to a compatible race of Xoo (Figure 6). Thus, Os Pti1a appears to negatively regulate plant resistance to both incompatible and compatible pathogens. Recent studies suggest that the R protein–mediated signaling pathway shares some components with the PAMP-triggered signaling pathway for basal resistance. Our results seem to be consistent with this notion. A possible explanation for our results would be that Pti1a lies at the point shared by both R protein– and PAMP receptor–mediated signaling pathways. Pti1a presumably suppresses defense signal transduction through modification of the common signaling components by its phosphorylation activity. Some results with regard to disease resistance, however, are apparently inconsistent with this conclusion. One of the inconsistencies is that no enhanced resistance was observed when the pti1a null mutant was challenged with incompatible M. grisea (containing avrPish). It could be that the strong Pish-mediated resistance masked the enhanced resistance in the pti1a mutant. Another inconsistency is that Os Pti1a-OE plants did not show reduced resistance to a compatible race of M. grisea. This is presumably because the strong pathogenicity of this pathogen overcomes Pti1a function.
Os Pti1a function Is Dependent on RAR1
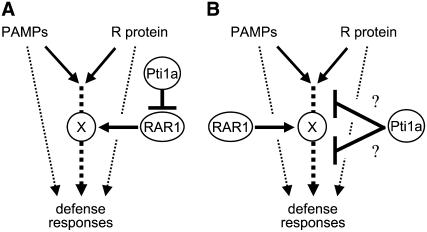
We have shown that the silencing of Os RAR1 cancels the lesion formation, PR gene expression, and acquired resistance against the compatible race of the blast fungus induced in the pti1a mutant, suggesting that Pti1a functions as a negative regulator of the rice defense signaling pathway genetically upstream of RAR1. In several plant species, RAR1 is required for the functioning of particular R proteins. In addition, rar1 mutation allowed enhanced susceptibility against the virulent bacterial strain P. syringae DC3000 in Arabidopsis and against the virulent fungus M. grisea in barley (Holt et al., 2005; Jarosch et al., 2005). Thus, RAR1 functions as a positive regulator of both basal resistance and gene-for-gene resistance. Indeed, in rice, RAR1-RNAi plants impaired basal resistance to blast fungus and bacterial blight (N.P. Thao, L. Chen, A. Nakashima, S. Hara, K. Umemura, A. Takahashi, K. Shirasu, T. Kawasaki, and K. Shimamoto, unpublished data). RAR1 is known to function as a molecular chaperone to stabilize NB-LRR protein in Arabidopsis, barley, and tobacco, although it is unclear whether Os RAR1 interacts with NB-LRR proteins in rice. On the basis of our observations, we propose two models for the defense signaling pathway in rice featuring Pti1a and RAR1. Since RAR1 functions as a molecular chaperone in other plant species, we postulate that Os RAR1 stabilizes an unknown protein, X, a presumptive essential component in the defense signaling pathway. In one model (Figure 8A), we propose that Os RAR1 positively regulates the signaling pathway through X, and Os Pti1a negatively regulates the signaling indirectly by repressing RAR1. In the second model (Figure 8B), Pti1a negatively regulates the defense signaling, which is dependent on RAR1 through the stabilization of X, by directly acting on the pathway upstream or downstream of X. In either model, upregulation of Pti1a and downregulation of RAR1 should result in the same outcome with respect to defense reactions. Our data are not necessarily consistent with this prediction: suppression of Os RAR1 in NB did not affect Pish-mediated resistance, whereas overexpression of Pti1a reduced the gene-for-gene resistance. However, given that RNAi-mediated downregulation is usually leaky, the downregulation of Os RAR1 expression in our Os RAR1-RNAi transformants could be less effective than the effects of Pti1a overexpression, irrespective of whether the negative regulation of the defense signaling pathway by Pti1a is indirect (Figure 8A) or direct (Figure 8B).
Figure 8.
Working Models of the Interplay of Pti1a and RAR1 in Defense Signaling in Rice.
PAMP-triggered basal resistance and R protein–mediated resistance mostly share common signaling pathways for induction of defense responses, including defense gene expression, accumulation of phytoalexin, and HR. We hypothesize that Os RAR1 positively regulates defense signal transduction required for both basal and R protein–mediated resistance through the unknown protein X. Os Pti1a negatively regulates the signaling pathway indirectly by repression of Os RAR1 (A) or by directly acting on the pathway upstream or downstream of X-Os RAR1 (B).
One possibility for constitutive activation of defense responses including cell death in the absence of Os Pti1a is explained by the guard hypothesis (Dangl and Jones, 2001). A knockout mutant of Arabidopsis RIN4, which is a negative regulator of basal resistance, is embryo-lethal, and the lethality was suppressed by elimination of the R gene RPS2 or delayed by a rar1 mutation leading to reduction of RPS2 protein accumulation, indicating that the elimination of RIN4 results in inappropriate RPS2-RAR1 activation (Mackey et al., 2003; Belkhadir et al., 2004). The relationship between Os Pti1a and X-RAR1 is reminiscent of that between RIN4 and RPS2-RAR1. A possible model based on this consideration would be that the unknown protein X is an NB-LRR protein, and the elimination of Os Pti1a invokes the NB-LRR-RAR1 complex, leading to activation of the signaling pathway that results in cell death and defense induction.
Defense Signal Transduction Mediated by Protein Phosphorylation
Protein phosphorylation appears to play a fundamental role in the early response of disease resistance. Some R proteins, including tomato Pto and rice Xa21, or the PAMP receptor FLS2 have protein kinase activity (Martin et al., 1993; Song et al., 1995; Gomez-Gomez and Boller, 2000). Calcium-dependent protein kinase and mitogen-activated protein kinase are well known as important regulators of the defense signaling cascade (Romeis, 2001). Furthermore, pharmacological analyses demonstrated that many protein kinase activities are required for the induction of both R protein– and PAMP receptor–mediated defense reactions (Lamb and Dixon, 1997; Takahashi et al., 1999). Recently, phosphor proteomics approaches have identified many proteins that undergo phosphorylation after treatment by elicitors or chemical inducers (Peck et al., 2001). However, in contrast with animals, only a few phosphorylation cascades have been characterized in plants. Therefore, identification of a protein kinase, or kinases, that phosphorylates Os Pti1a could provide some very useful clues to understanding phosphorylation-mediated signaling in the negative regulation of defense. The Thr residue at 233 (Thr-233) in Sl Pti1, which is the major phosphorylation site of Pto (Sessa et al., 2000), is conserved at the corresponding positions in both Os Pti1a (Thr-233) and Os Pti1b (Thr-236). This may imply that an as yet unidentified rice ortholog of Pto may be an upstream protein kinase that phosphorylates Pti1a.
Genetic Screening in Rice
Using transgenic tobacco, Sl Pti1 was shown to be a positive regulator of HR induction triggered by Pto–avrPto interaction (Zhou et al., 1995). However, there are no loss-of-function data to support the involvement of Sl Pti1 in Pto-dependent disease resistance. This may be explained by its functional redundancy because Sl Pti1 appears to be a member of a gene family that encodes a group of closely related protein kinases (Mysore et al., 2002). Arabidopsis has at least five Pti1 homologs in its genome. Rice has only two highly conserved Pti1 isoforms, Pti1a and Pti1b (Figure 3). Pti1a alone was isolated in our genetic screens despite the apparent genetic redundancy likely because of the extremely low expression level of Pti1b compared with Pti1a (Figure 4). This case illustrates the merits of using rice instead of Arabidopsis in a genetic approach to find novel proteins in a conserved signal transduction pathway when redundancy could be a problem in Arabidopsis and other plant species. Interestingly, expression of Sl Pti1 complements the Os pti1a phenotype (Figure 5), indicating that Sl Pti1 acts as a negative regulator of the HR response in rice, while it behaves as a positive regulator in tobacco. Therefore, although protein function is conserved beyond plant species, the signal cascade leading to HR downstream of Pti1 may have evolved differently in rice and tomato.
Despite the passage of more than a decade since the first molecular cloning of R proteins, there is very little definitive understanding of how they transduce pathogen recognition signals or activate defense responses, including HR. Os Pti1a functions as a negative regulator of defense responses associated with HR downstream of the R protein and PAMP signaling. Therefore, further understanding of Os Pti1a function should lead to a fuller understanding of the molecular mechanisms of HR induction and defense signaling pathways.
METHODS
Plant and Pathogen Materials
The pti1a mutant is derived from rice (Oryza sativa) Japonicum cultivar NB mutant lines induced by insertion of the rice endogenous retrotransposon Tos17 (Hirochika, 2001). NB carries the blast resistance genes Pish and Pi19. Strains of Magnaporthe grisea, Kyu89-246 (MAFF101506; race 003.0) as compatible and Kyu77-07A (avrPish; Race 102.0) and CHNOS58-3-1 (avr19; Race 000.0) as incompatible races, were used in this experiment. Thus, NB is resistant to Kyu89-246 and CHNOS58-3-1 and susceptible to Kyu-77-07A. M. grisea was grown on oatmeal agar medium (30 g/L oatmeal, 5 g/L sucrose, and 16 g/L agar) at 22°C. Seedlings were inoculated at the four- to six-leaf stage by spraying an aqueous spore suspension containing 105 to 1.5 × 105 spores per mL to runoff. Inoculated seedlings were kept in a dark chamber with a moisture-saturated atmosphere at 24°C for 20 h and then maintained at 27°C and 70 to 80% relative humidity in a greenhouse. Disease development was monitored 1 week after inoculation. Lesion size per each leaf was measured and calculated using a digital microscope VHX500 system (KEYENCE). Methods for the punch infection of the leaf blade with the blast fungus have been described (Takahashi et al., 1999). Bacterial blight inoculation experiments were performed with the Japanese Xoo race 1 using a scissors-dip method (Kauffman et al., 1973). Lesion development was scored on rice leaves 21 d after inoculation by measuring margin progression with a ruler.
RNA Analysis
Total RNA was isolated from rice seedling roots, leaves, or panicles as described previously (Agrawal et al., 2001), separated on 1.2% (w/v) formaldehyde-denaturing agarose gels, and blotted onto nylon membranes (Hybond N+; Amersham). The cDNA fragments corresponding to Os Pti1a, Pti1b, PR1b, PR5, PR10a, and PAL were amplified by PCR from wild-type leaf cDNAs using gene-specific primers (Takahashi et al., 1999; Agrawal et al., 2000). The cDNA fragment corresponding to Sl Pti1 was amplified by PCR from tomato leaf cDNA using the specific primers L (5′-CCCACACTTTCAAGAAGGTTAGAATC-3′) and R (5′-CACAAATTCACGATCCTCTTGTG-3′). Primers for RT-PCR analyses were AOL20 (5′-GACTCTAGTAAGCAGCCAGACC-3′) and cDNA875R (5′-AGGTGCATGATATCCAAAGG-3′) for Os Pti1a, AOL33 (5′-TCATGATGGCATGAAACAGTGGAG-3′) and AOL34 (5′-GGTGGAACTGCAGGCTTCTCAAC-3′) for Os RAR1, AOL37 (5′-AGGTATCCAAGCTGGCCATTG-3′) and AOL38 (5′-TATGGACCGTGGACCTGTTTAC-3′) for PR1b, and Os Act1U (5′-TCCATCTTGGCATCTCTCAG-3′) and Os Act1L (5′-GTACCCGCATCAGGCATCTG-3′) for rice Actin.
Phytoalexin Measurement
For measuring the accumulation of the phytoalexin momilactone A, leaves (the middle portion only) from three to four individual plants were used. Leaf samples were harvested from wild-type plants and from mutant plants before and after lesion formation. Harvested leaves were immediately frozen in liquid nitrogen to prevent touch- or wound-induced accumulation of phytoalexins. Quantification of momilactone A was performed as described previously (Takahashi et al., 1999). Briefly, leaves were cut into small pieces, transferred to a glass test tube containing 5 mL of 80% aqueous methanol, and boiled for 5 min. Three microliters of the crude extract was injected onto an HPLC and analyzed by liquid chromatography–tandem mass spectrometry (Ibid).
Isolation of Tos17 Insertion Sites and Reverse Genetic Analysis
Sequences flanking Tos17 insertions were amplified by thermal asymmetric interlaced PCR as previously described (Yamazaki et al., 2001). For reverse genetic analyses, Tos17-specific primers were used in combination with the Os Pti1a and Os Pti1b gene-specific primers. Two Tos17 (T17F-1 [5′-ACCACTTCAGAGATTGTGTGGTTGC-3′] and T17R-1 [5′-CAGCAACGATGTAGATGGTCAAGC-3′]) and two Os Pti1a– or two Os Pti1b–specific primers were used in all possible combinations for PCR amplification of genomic DNA from each pooled DNA sample.
Sequence Analysis
Multiple sequence alignments were produced with a Web-based version of ClustalW (http://crick.genes.nig.ac.jp/homology/clustalw-e.shtml) using default settings (Matrix = blossom; GAPOPEN = 0, GAPEXT = 0, GAPDIST = 8, and MAXDIV = 40). The phylogenetic tree was calculated using the neighbor-joining method and bootstrap analysis (1000 replicates) using PHYLIP via the same website and visualized with Treeviewer version 1.6.6 (http://taxonomy.zoology.gla.ac.uk/rod/rod.html).
Rice Transformation
To overexpress Os Pti1a and Sl Pti1 cDNA, the coding sequences were cloned into the Ti-based vector pPZP2Ha3(+) downstream of the cauliflower mosaic virus 35S promoter, and Agrobacterium tumefaciens–mediated transformation of rice callus was performed according to a published protocol (Hiei et al., 1994; Fuse et al., 2001). Plants regenerated from hygromycin-resistant calluses were grown in an isolated greenhouse. For the complementation experiment, seeds harvested from the heterozygous Os pti1a (+/−) plant were used for callus induction, and the genotypes of regenerated plants were determined by PCR using PA1326 (5′-TGGAAATCTCCGTGTCCTTGC-3′), PA1327 (5′-ACTCCAGGCCTTTTGCTGCC-3′), and PA0234 (5′-ACCACTTCAGAGATTGTGTGGTTGC-3′). To suppress Os RAR1 expression, a cDNA fragment amplified by PCR using two primers, Os RAR1-F (5′-TCTGAGTGAGCCTAGGGTTTG-3′) and Os RAR1-R (5′-GACCGAAGTCTCCACACACA-3′), was fused in reverse orientation, and an unrelated fragment of the Escherichia coli β-glucuronidase gene was inserted as a linker (Miki and Shimamoto, 2004). This construct was then fused with the maize (Zea mays) Ubq1 promoter and introduced as above. More than three independent transgenic lines for each experiment were produced, and the expression level was confirmed by RNA or protein gel blot analysis.
Antibody Production and Protein Gel Blot Analysis
Polyclonal anti-Os Pti1a antibody was generated in rabbits using the peptide including Os Pti1a amino acids 11 to 24 as an antigen and antigen-purified before use for protein gel blot analysis. Total protein extracts were prepared from leaves in 100 mM Tris-HCl, pH 8.5, 4% (w/v) SDS, 20% (w/v) glycerol, and 2% (v/v) 2-mercaptoethanol and separated on 10% (w/v) SDS-PAGE gels.
Expression of Proteins and in Vitro Kinase Assay
Os Pti1a and its mutagenized form (K96N) were expressed as fusion proteins with an N terminus poly-histidine tag using a bacterial expression system (Invitrogen) following the supplier's instructions. Proteins were purified by immobilized metal ion affinity chromatography and applied for autophosphorylation assay as described (Zhou et al., 1995) with small modifications (addition of NaCl to the reaction buffer to a final concentration of 100 mM). Proteins were fractionated by SDS-PAGE and stained with Coomassie Brilliant Blue, and the 32P-labeled fractions were detected by autoradiography.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: AK104870 (Os Pti1a), AK065231 (Os Pti1b), U28007 (Sl Pti1), AK111881 (Os RAR1), U89895 (Os PR1b), X68197 (Os PR5), D38170 (Os PR10a), and X87946 (Os PAL).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Effect of Overexpression of Os Pti1a on Resistance against M. grisea (avr19).
Supplemental Figure 2. Suppression of Os RAR1 Expression Does Not Affect Lesion Formation in cdr Mutants.
Supplementary Material
Acknowledgments
We thank Morifumi Hasegawa for measuring momilactone A and Mayuko Yamazaki for her technical assistance. We gratefully acknowledge critical comments from Hiroshi Taktsuji. This work was supported by a grant from the Ministry of Agriculture, Forestry, and Fisheries of Japan (Green Technology Project IP-4002).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Hirohiko Hirochika (hirohiko@nias.affrc.go.jp).
Online version contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Agrawal, G.K., Rakwal, R., and Jwa, N.S. (2000). Rice (Oryza sativa L.) OsPR1b gene is phytohormonally regulated in close interaction with light signals. Biochem. Biophys. Res. Commun. 278 290–298. [DOI] [PubMed] [Google Scholar]
- Agrawal, G.K., Yamazaki, M., Kobayashi, M., Hirochika, R., Miyao, A., and Hirochika, H. (2001). Screening of the rice viviparous mutants generated by endogenous retrotransposon Tos17 insertion. Tagging of a zeaxanthin epoxidase gene and a novel ostatc gene. Plant Physiol. 125 1248–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azevedo, C., Sadanandom, A., Kitagawa, K., Freialdenhoven, A., Shirasu, K., and Schulze-Lefert, P. (2002). The RAR1 interactor SGT1, an essential component of R gene-triggered disease resistance. Science 295 2073–2076. [DOI] [PubMed] [Google Scholar]
- Belkhadir, Y., Nimchuk, Z., Hubert, D.A., Mackey, D., and Dangl, J.L. (2004). Arabidopsis RIN4 negatively regulates disease resistance mediated by RPS2 and RPM1 downstream or independent of the NDR1 signal modulator and is not required for the virulence functions of bacterial type III effectors AvrRpt2 or AvrRpm1. Plant Cell 16 2822–2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartwright, D., Langcake, P., Pryce, D.P., Leworthy, D.P., and Ride, J.P. (1977). Chemical activation of host defence mechanisms as a basis for crop protection. Nature 361 153–156. [Google Scholar]
- Chisholm, S.T., Coaker, G., Day, B., and Staskawicz, B.J. (2006). Host-microbe interactions: Shaping the evolution of the plant immune response. Cell 124 803–814. [DOI] [PubMed] [Google Scholar]
- Dangl, J.L., and Jones, J.D. (2001). Plant pathogens and integrated defence responses to infection. Nature 411 826–833. [DOI] [PubMed] [Google Scholar]
- Durrant, W.E., and Dong, X. (2004). Systemic acquired resistance. Annu. Rev. Phytopathol. 42 185–209. [DOI] [PubMed] [Google Scholar]
- Freialdenhoven, A., Scherag, B., Hollricher, K., Collinge, D.B., Thordal-Christensen, H., and Schulze-Lefert, P. (1994). Nar-1 and Nar-2, two loci required for Mla12-specified race-specific resistance to powdery mildew in barley. Plant Cell 6 983–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuse, T., Sasaki, T., and Yano, M. (2001). Ti-plasmid vectors useful for functional analysis of rice genes. Plant Biotechnol. 18 219–222. [Google Scholar]
- Gomez-Gomez, L., and Boller, T. (2000). FLS2: An LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol. Cell 5 1003–1011. [DOI] [PubMed] [Google Scholar]
- Greenberg, J.T., and Yao, N. (2004). The role and regulation of programmed cell death in plant-pathogen interactions. Cell. Microbiol. 6 201–211. [DOI] [PubMed] [Google Scholar]
- Hammond-Kosack, K.E., and Parker, J.E. (2003). Deciphering plant-pathogen communication: Fresh perspectives for molecular resistance breeding. Curr. Opin. Biotechnol. 14 177–193. [DOI] [PubMed] [Google Scholar]
- Hayashi, N., Ando, I., and Imbe, T. (1998). Identification of a new resistance gene to a Chinese blast fungus isolate in the japanese rice cultivar aichi asahi. Phytopathology 88 822–827. [DOI] [PubMed] [Google Scholar]
- Hiei, Y., Ohta, S., Komari, T., and Kumashiro, T. (1994). Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 6 271–282. [DOI] [PubMed] [Google Scholar]
- Hirochika, H. (2001). Contribution of the Tos17 retrotransposon to rice functional genomics. Curr. Opin. Plant Biol. 4 118–122. [DOI] [PubMed] [Google Scholar]
- Hirochika, H., Guiderdoni, E., An, G., Hsing, Y.I., Eun, M.Y., Han, C.D., Upadhyaya, N., Ramachandran, S., Zhang, Q., Pereira, A., Sundaresan, V., and Leung, H. (2004). Rice mutant resources for gene discovery. Plant Mol. Biol. 54 325–334. [DOI] [PubMed] [Google Scholar]
- Holt III, B.F., Belkhadir, Y., and Dangl, J.L. (2005). Antagonistic control of disease resistance protein stability in the plant immune system. Science 309 929–932. [DOI] [PubMed] [Google Scholar]
- Imbe, T., and Matsumoto, S. (1985). Inheritance of resistance of rice varieties to the blast fungus strains virulent to the variety “Reiho”. Jap. J. Breed 35 332–339. [Google Scholar]
- Jarosch, B., Collins, N.C., Zellerhoff, N., and Schaffrath, U. (2005). RAR1, ROR1, and the actin cytoskeleton contribute to basal resistance to Magnaporthe grisea in barley. Mol. Plant Microbe Interact. 18 397–404. [DOI] [PubMed] [Google Scholar]
- Jones, D.A., and Takemoto, D. (2004). Plant innate immunity - Direct and indirect recognition of general and specific pathogen-associated molecules. Curr. Opin. Immunol. 16 48–62. [DOI] [PubMed] [Google Scholar]
- Jones, J.D., and Dangl, J.L. (2006). The plant immune system. Nature 444 323–329. [DOI] [PubMed] [Google Scholar]
- Kauffman, H.E., Reddy, A.P.K., Hsieh, S.P.Y., and Merca, S.D. (1973). An improved technique for evaluating resistance to rice varieties of Xanthomonas oryzae pv. oryzae. Plant Dis. Rep. 57 537–541. [Google Scholar]
- Kim, M.G., da Cunha, L., McFall, A.J., Belkhadir, Y., DebRoy, S., Dangl, J.L., and Mackey, D. (2005). Two Pseudomonas syringae type III effectors inhibit RIN4-regulated basal defense in Arabidopsis. Cell 121 749–759. [DOI] [PubMed] [Google Scholar]
- Lamb, C., and Dixon, R.A. (1997). The Oxidative burst in plant disease resistance. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48 251–275. [DOI] [PubMed] [Google Scholar]
- Liu, Y., Schiff, M., Marathe, R., and Dinesh-Kumar, S.P. (2002). Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for N-mediated resistance to tobacco mosaic virus. Plant J. 30 415–429. [DOI] [PubMed] [Google Scholar]
- Liu, Y.G., Mitsukawa, N., Oosumi, T., and Whittier, R.F. (1995). Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J. 8 457–463. [DOI] [PubMed] [Google Scholar]
- Mackey, D., Belkhadir, Y., Alonso, J.M., Ecker, J.R., and Dangl, J.L. (2003). Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2-mediated resistance. Cell 112 379–389. [DOI] [PubMed] [Google Scholar]
- Martin, G.B., Brommonschenkel, S.H., Chunwongse, J., Frary, A., Ganal, M.W., Spivey, R., Wu, T., Earle, E.D., and Tanksley, S.D. (1993). Map-based cloning of a protein kinase gene conferring disease resistance in tomato. Science 262 1432–1436. [DOI] [PubMed] [Google Scholar]
- Miki, D., and Shimamoto, K. (2004). Simple RNAi vectors for stable and transient suppression of gene function in rice. Plant Cell Physiol. 45 490–495. [DOI] [PubMed] [Google Scholar]
- Muskett, P.R., Kahn, K., Austin, M.J., Moisan, L.J., Sadanandom, A., Shirasu, K., Jones, J.D., and Parker, J.E. (2002). Arabidopsis RAR1 exerts rate-limiting control of R gene-mediated defenses against multiple pathogens. Plant Cell 14 979–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mysore, K.S., Crasta, O.R., Tuori, R.P., Folkerts, O., Swirsky, P.B., and Martin, G.B. (2002). Comprehensive transcript profiling of Pto- and Prf-mediated host defense responses to infection by Pseudomonas syringae pv. tomato. Plant J. 32 299–315. [DOI] [PubMed] [Google Scholar]
- Nimchuk, Z., Eulgem, T., Holt III, B.F., and Dangl, J.L. (2003). Recognition and response in the plant immune system. Annu. Rev. Genet. 37 579–609. [DOI] [PubMed] [Google Scholar]
- Nurnberger, T., Brunner, F., Kemmerling, B., and Piater, L. (2004). Innate immunity in plants and animals: Striking similarities and obvious differences. Immunol. Rev. 198 249–266. [DOI] [PubMed] [Google Scholar]
- Peart, J.R., et al. (2002). Ubiquitin ligase-associated protein SGT1 is required for host and nonhost disease resistance in plants. Proc. Natl. Acad. Sci. USA 99 10865–10869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peck, S.C., Nuhse, T.S., Hess, D., Iglesias, A., Meins, F., and Boller, T. (2001). Directed proteomics identifies a plant-specific protein rapidly phosphorylated in response to bacterial and fungal elicitors. Plant Cell 13 1467–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedley, K.F., and Martin, G.B. (2003). Molecular basis of Pto-mediated resistance to bacterial speck disease in tomato. Annu. Rev. Phytopathol. 41 215–243. [DOI] [PubMed] [Google Scholar]
- Piffanelli, P., Ramsay, L., Waugh, R., Benabdelmouna, A., D'Hont, A., Hollricher, K., Jorgensen, J.H., Schulze-Lefert, P., and Panstruga, R. (2004). A barley cultivation-associated polymorphism conveys resistance to powdery mildew. Nature 430 887–891. [DOI] [PubMed] [Google Scholar]
- Romeis, T. (2001). Protein kinases in the plant defence response. Curr. Opin. Plant Biol. 4 407–414. [DOI] [PubMed] [Google Scholar]
- Sessa, G., D'Ascenzo, M., and Martin, G.B. (2000). The major site of the pti1 kinase phosphorylated by the pto kinase is located in the activation domain and is required for pto-pti1 physical interaction. Eur. J. Biochem. 267 171–178. [DOI] [PubMed] [Google Scholar]
- Shang, Y., Li, X., Cui, H., He, P., Thilmony, R., Chintamanani, S., Zwiesler-Vollick, J., Gopalan, S., Tang, X., and Zhou, J.M. (2006). RAR1, a central player in plant immunity, is targeted by Pseudomonas syringae effector AvrB. Proc. Natl. Acad. Sci. USA 103 19200–19205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirasu, K., Lahaye, T., Tan, M.W., Zhou, F., Azevedo, C., and Schulze-Lefert, P. (1999). A novel class of eukaryotic zinc-binding proteins is required for disease resistance signaling in barley and development in C. elegans. Cell 99 355–366. [DOI] [PubMed] [Google Scholar]
- Shirasu, K., and Schulze-Lefert, P. (2003). Complex formation, promiscuity and multi-functionality: Protein interactions in disease-resistance pathways. Trends Plant Sci. 8 252–258. [DOI] [PubMed] [Google Scholar]
- Song, W.Y., Wang, G.L., Chen, L.L., Kim, H.S., Pi, L.Y., Holsten, T., Gardner, J., Wang, B., Zhai, W.X., Zhu, L.H., Fauquet, C., and Ronald, P. (1995). A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science 270 1804–1806. [DOI] [PubMed] [Google Scholar]
- Takahashi, A., Casais, C., Ichimura, K., and Shirasu, K. (2003). HSP90 interacts with RAR1 and SGT1 and is essential for RPS2-mediated disease resistance in Arabidopsis. Proc. Natl. Acad. Sci. USA 100 11777–11782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, A., Kawasaki, T., Henmi, K., Shi, I.K., Kodama, O., Satoh, H., and Shimamoto, K. (1999). Lesion mimic mutants of rice with alterations in early signaling events of defense. Plant J. 17 535–545. [DOI] [PubMed] [Google Scholar]
- Tornero, P., Merritt, P., Sadanandom, A., Shirasu, K., Innes, R.W., and Dangl, J.L. (2002). RAR1 and NDR1 contribute quantitatively to disease resistance in Arabidopsis, and their relative contributions are dependent on the R gene assayed. Plant Cell 14 1005–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki, M., Tsugawa, H., Miyao, A., Yano, M., Wu, J., Yamamoto, S., Matsumoto, T., Sasaki, T., and Hirochika, H. (2001). The rice retrotransposon Tos17 prefers low-copy-number sequences as integration targets. Mol. Genet. Genomics 265 336–344. [DOI] [PubMed] [Google Scholar]
- Zhou, J., Loh, Y.T., Bressan, R.A., and Martin, G.B. (1995). The tomato gene Pti1 encodes a serine/threonine kinase that is phosphorylated by Pto and is involved in the hypersensitive response. Cell 83 925–935. [DOI] [PubMed] [Google Scholar]
- Zipfel, C., and Felix, G. (2005). Plants and animals: A different taste for microbes? Curr. Opin. Plant Biol. 8 353–360. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.