Abstract
Naturally existing variation in the eukaryotic translation initiation factor 4E (eIF4E) homolog encoded at the pvr1 locus in Capsicum results in recessively inherited resistance against several potyviruses. Previously reported data indicate that the physical interaction between Capsicum-eIF4E and the viral genome-linked protein (VPg) is required for the viral infection in the Capsicum-Tobacco etch virus (TEV) pathosystem. In this study, the potential structural role(s) of natural variation in the eIF4E protein encoded by recessive resistance alleles and their biological consequences have been assessed. Using high-resolution three-dimensional structural models based on the available crystallographic structures of eIF4E, we show that the amino acid substitution G107R, found in many recessive plant virus resistance genes encoding eIF4E, is predicted to result in a substantial modification in the protein binding pocket. The G107R change was shown to not only be responsible for the interruption of VPg binding in planta but also for the loss of cap binding ability in vitro, the principal function of eIF4E in the host. Overexpression of the Capsicum-eIF4E protein containing the G107R amino acid substitution in Solanum lycopersicum indicated that this polymorphism alone is sufficient for the acquisition of resistance against several TEV strains.
INTRODUCTION
Precise molecular interactions between host and pathogen occur when disease develops or successful host defense is initiated. At one end of this spectrum, these interactions, which may be direct or indirect, allow plants to activate a defense response that is often mediated by resistance genes (Dangl and Jones, 1998, 2001; Nimchuk et al., 2003). For disease development to occur, on the other hand, the invading pathogen must establish a specific molecular relationship with the host plant that provides an environment supportive of pathogen infection and its life cycle.
Because viruses are obligate intracellular parasites lacking the components necessary for their own independent survival, they rely upon an array of host factors to support their life cycle. The identification of host factors required for viral disease has provided useful information concerning virus infection. For instance, it has been observed that geminiviruses use host factors, such as the proliferating cell nuclear agent, a DNA polymerase accessory factor, NAC domain protein (SINAC1), a transcription factor, and plant homologs of retinoblastoma protein, to reprogram the host's cell cycle to allow replication of their own genome (Gutierrez, 2000; Gutierrez et al., 2004; Rojas et al., 2005). Likewise, several host factors for Tomato bushy stunt virus, a tombusvirus, and Brome mosaic virus, a bromovirus, have been thoroughly investigated using yeast as an artificial host system (Janda and Ahlquist, 1993; Duggal and Hall, 1995; Ishikawa et al., 1997; Noueiry and Ahlquist, 2003; Panavas et al., 2005; Mas et al., 2006; Serviene et al., 2006).
The physical interaction between host eukaryotic initiation factor eIF4E or eIF(iso)4E and the viral genome-linked protein (VPg) is critical for viral infection by several members of the genus potyvirus (Wittmann et al., 1997; Leonard et al., 2000; Grzela et al., 2006; Miyoshi et al., 2006; Robaglia and Caranta, 2006). The major role of eIF4E in the host cell is initiating protein translation by allowing recognition and interaction with the cap structure of cellular mRNA. However, it has been shown that the viral protein VPg can interfere with cap binding ability of eIF4E or its isoform eIF(iso)4E, which likely is responsible for the interruption of the formation of the translation initiation complex (Miyoshi et al., 2006). Additionally, a number of recent studies have demonstrated that eIF4G, which functions as an adapter connecting the translation initiation complex with the 40S ribosomal subunit, and the leader sequence of the Tobacco etch virus (TEV) RNA genome are also involved in the formation of the complex including eIF4E and VPg (Khan et al., 2006; Michon et al., 2006; Ray et al., 2006; Nicaise et al., 2007), which further demonstrates the involvement of the host translation initiation machinery in the initial steps of viral infection. Interestingly, the involvement of host translation initiation machinery during the viral infection process is not unique to the plant/potyvirus pathosystem. Several animal viruses, such as adenovirus, influenza, and poliovirus, are known to manipulate host translation machinery by inactivating or interrupting certain translation initiation processes (Sonenberg, 1987; Sonenberg and Pelletier, 1989; Mathews, 1990; Garfinkel and Katze, 1992; Cuesta et al., 2000; Gale et al., 2000).
Because viruses rely upon host factors for their own survival during the infection process, interruption of infection could result when a host factor is modified or eliminated such that infection could no longer progress (Kasschau and Carrington, 1998; Carrington et al., 2001; Baulcombe, 2004; Kang et al., 2005b; Soosaar et al., 2005). The efficacy of this resistance, which typically manifests as recessive inheritence, is evidenced by the prominence of recessive resistance to plant viruses relative to that of other pathogen classes (Provvidenti and Hampton, 1992; Keller et al., 1998; Robaglia and Caranta, 2006). This defense strategy appears to be particularly widely observed at the eIF4E locus where resistance to multiple RNA phytopathogenic viruses occurs in numerous plant families, including pvr1 in Capsicum (Ruffel et al., 2002; Kang et al., 2005a), mo1 in lettuce (Lactuca sativa; Nicaise et al., 2003), sbm1 in pea (Pisum sativum; Gao et al., 2004), pot-1 in tomato (Solanum lycopersicum; Ruffel et al., 2005), rym4/5 in barley (Hordeum vulgare; Stein et al., 2005), and nsv in melon (Cucumis melo; Nieto et al., 2006). In addition to these naturally existing genes, lsp1 and cum1 resistance alleles created via mutagenesis in Arabidopsis thaliana also encode eIF4E (Lellis et al., 2002; Yoshii et al., 2004). Thus, it is clear that genetic resistance against many viruses may arise when a plant expresses a gene encoding an altered eIF4E that interrupts its role in viral infection. Previous studies have shown that the physical interaction between eIF4E and VPg is correlated with viral infection (Wittmann et al., 1997; Lellis et al., 2002; Kang et al., 2005a). In the case of recessively inherited resistance against potyviruses in Capsicum, natural variation in eIF4E encoded by recessive resistance alleles are responsible for nonconservative amino acid changes that disrupt interaction with TEV VPg (Kang et al., 2005a).
This article aims to address the functional significance of natural amino acid variation in recessive resistance alleles with respect to the disease resistance phenotype. Mutational dissection of eIF4E defined the potential structural role(s) of each substitution in recessive resistance assayed at the organismal and at the protein levels. Examination of the eIF4E proteins containing dissected alterations allowed us to determine the significance of each substitution with respect to changes in affinity with their viral counterpart VPg in planta. Biological function of critical amino acid changes was determined by assessing the phenotypic outcome of transgenic tomato plants expressing an eIF4E allele containing a single substitution. Based on these results, we conclude that a Gly-to-Arg change at position 107 (G107R) in the eIF4E protein encoded by the pvr1 allele defines the most significant position for host resistance to potyviruses. The amino acid change at this position is observed in multiple recessive potyvirus resistance genes from diverse plants, including pepper (Capsicum annuum), lettuce, and pea. Elucidation of the importance of this particular substitution as the molecular basis for the resistance phenotype opens the way to further detailed study of the resistance phenotype, the range of viral genotypes controlled, and the durability of resistance in nature.
RESULTS
Potyvirus Resistance Is Controlled by Amino Acid Changes in eIF4E Encoded at the pvr1 Locus
Single nucleotide polymorphisms within the coding region of eIF4E encoded by multiple recessive resistance alleles at the pvr1 locus, pvr1, pvr11, and pvr12, compared with the susceptible allele Pvr1+ result in nonconservative amino acid changes in the eIF4E protein. These single nucleotide polymorphisms causing amino acid changes in the eIF4E protein determine both the range of potyviral isolates controlled by the resistance alleles and the degree to which viral titer and symptoms are reduced (Kang et al., 2005a). The expression level of eIF4E protein was examined in various pepper genotypes before and after inoculation of the highly aphid-transmissable (HAT) strain of TEV to determine whether potyvirus resistance is a result of differences in protein expression or accumulation. There were no obvious correlations between the eIF4E protein level and viral resistance in all pepper genotypes tested in this study (Figure 1A). Accumulation of TEV coat protein in susceptible genotypes was confirmed by an immunoblot using an antibody for TEV coat protein. Additionally, no differences in total RNA level were observed among genotypes included in this study (data not shown). These results support the prevailing hypothesis that phenotypic differences in potyviral infection among these genotypes are determined by the amino acid changes themselves, rather than by other components regulating expression or accumulation of eIF4E protein.
Figure 1.
Amino Acid Changes in eIF4E Protein Encoded at the Capsicum pvr1 Locus Are Responsible for Gain of Resistance.
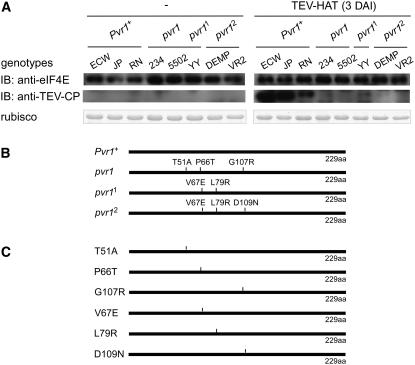
(A) Pepper genotypes homozygous at the pvr1 locus are expressing eIF4E protein at similar levels. The expression level of eIF4E protein encoded by the pvr1 locus was examined in various C. annuum and C. chinense genotypes before and after TEV-HAT inoculation. Crude protein extracts obtained from pepper leaf tissue were loaded on a 12% SDS-PAGE gel. Immunoblot assay was performed using antibodies for Capsicum-eIF4E (top) and coat protein of TEV (middle). The amount of the loaded protein extract was shown by Coomassie blue staining (bottom). Rubisco, ribulose-1,5-bisphosphate carboxylase/oxygenase.
(B) Diagram indicating amino acid changes naturally occurring in the resistance alleles at the pvr1 locus. eIF4E-pvr1 contains T51A, P66T, and G107R amino acid substitutions; eIF4E-pvr11 contains V67E and L79R amino acid substitutions; and eIF4E-pvr12 contains V67E, L79R, and D109N amino acid substitutions compared with eIF4E-Pvr1+.
(C) Diagram showing eIF4Es containing individual substitution separately: eIF4E-T51A, eIF4E-P66T, eIF4E-G107R, eIF4E-V67E, eIF4E-L79R, and eIF4E-D109N. Site-directed mutagenesis was performed to create single point mutations in eIF4E-Pvr1+.
The naturally occurring amino acid substitutions in eIF4E encoded by resistance alleles at the pvr1 locus are indicated in Figure 1B. The eIF4E protein encoded by the pvr1 allele (eIF4E-pvr1) contains amino acid changes T51A, P66T, and G107R, eIF4E-pvr11 contains amino acid changes V67E and L79R, and eIF4E-pvr12 contains amino acid changes V67E, L79R, and D109N compared with eIF4E-Pvr1+ (Ruffel et al., 2002; Kang et al., 2005a). To understand the biochemical effect of each amino acid substitution in eIF4E, we generated novel alleles of eIF4E, which contain each amino acid substitution separately, as shown in Figure 1C. The eIF4E constructs containing each substitution, eIF4E-T51A, eIF4E-P66T, eIF4E-G107R, eIF4E-V67E, eIF4E-L79R, and eIF4E-D109N, were applied in further assays.
The G107R Amino Acid Change in the pvr1 Allele Is Responsible for Abolishing eIF4E–VPg Interaction in Yeast
The physical interaction between eIF4E encoded by resistance alleles at the pvr1 locus and VPg from different TEV strains has been investigated previously using both yeast two-hybrid and glutathione S-transferase pull-down assays (Kang et al., 2005a). eIF4E encoded by the susceptible allele (eIF4E-Pvr1+) interacted strongly with VPg proteins from different TEV strains. However, VPg interaction was not detected for eIF4E-pvr1, eIF4E-pvr11, or eIF4E-pvr12, which implies that the T51A, P66T, and G107R amino acid changes in eIF4E-pvr1 as well as the V67E and L79R amino acid changes in both eIF4E-pvr11 and eIF4E-pvr12 are responsible for the interruption of the physical interaction between eIF4E and VPg in yeast.
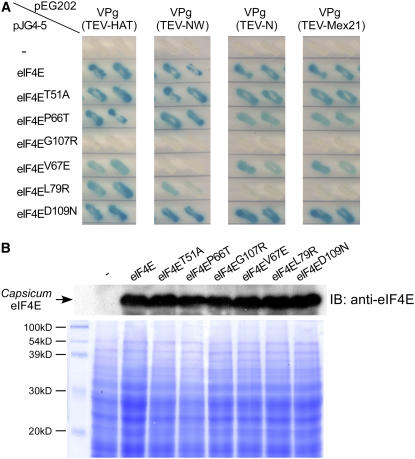
To examine the effect of each amino acid change on the physical interaction between eIF4E and VPg, yeast two-hybrid assays using eIF4E-T51A, eIF4E-P66T, eIF4E-G107R, eIF4E-V67E, eIF4E-L79R, and eIF4E-D109N with the VPg protein encoded by four different TEV strains: TEV-HAT, TEV-NW, TEV-N, and TEV-Mex21 (see Methods), were performed (Figure 2A). eIF4E-Pvr1+ interacted very strongly with all VPg proteins from four different TEV strains; this interaction served as a positive control for each TEV strain. The proteins eIF4E-T51A, eIF4E-P66T, and eIF4E-D109N all interacted with VPg, showing no visible difference from results obtained in the positive control for each TEV strain. By contrast, eIF4E-V67E and eIF4E-L79R, each of which contain a single substitution observed in both eIF4E-pvr11 and eIF4E-pvr12, showed inconsistent interaction with VPg protein from four TEV strains. In the case of eIF4E-V67E, the interaction with VPg from TEV-NW was noticeably reduced, but the weaker interaction was less evident with VPg proteins from the other TEV strains. eIF4E–VPg interaction was greatly diminished for eIF4E-L79R when TEV-N and TEV-Mex21 were used. These results suggest that the complete loss of VPg binding ability of eIF4E encoded by the pvr11 and pvr12 alleles described in the previous study (Kang et al., 2005a) is the result of an additive effect of the V67E and L79R changes. It is striking that eIF4E-G107R existing in eIF4E-pvr1, which displays the broadest resistance spectrum among resistance alleles at the pvr1 locus, did not interact with VPg proteins from any of the four different TEV strains. Together, these results indicate that the loss of VPg binding ability of eIF4E-pvr1 is caused by the single change G107R because the two other changes in eIF4E-pvr1, T51A and P66T, did not affect eIF4E–VPg interaction. Reliable expression of eIF4E protein for each yeast cell used in yeast two-hybrid assays was confirmed by immunoblot analysis (Figure 2B).
Figure 2.
Interaction between eIF4E Protein Containing a Single Amino Acid Change and VPg from Four Different TEV Strains Was Examined via Yeast Two-Hybrid Assay.
(A) X-galactosidase assay of yeast two-hybrid interaction between TEV-VPg and eIF4Es. Bait plasmid pEG202 was used to express the fusion protein VPg from TEV-HAT, TEV-NW, TEV-N, or TEV-Mex21, while the prey plasmid pJG4-5 was used to express eIF4E containing T51A, P66T, G107R, V67E, L79R, or D109N. The empty vector pJG4-5 served as a negative interaction control. Yeast cells containing known interactors pEG202:VPg and pJG4-5:eIF4E-Pvr1+ served as a positive interaction control.
(B) Expression of eIF4E proteins in yeast cells was examined via immunoblot (IB) assay. Yeast cells containing empty vector (lane 1) or eIF4E fusion gene in pJG4-5 vector (lanes 2 to 8) were grown on selection medium lacking Leu and using galactose as a carbon source. Proteins were fractionated on 12% SDS-polyacrylamide gels and immunoblotted with antibody for Capsicum-eIF4E (top panel). The eIF4E fusion proteins ∼48 kD were detected in all yeast cells containing various eIF4E genes, as indicated by the arrow, but not in the cells containing empty vector. Coomassie blue–stained gel showing equal amounts of protein extract loaded on the gel (bottom panel).
The G107R Amino Acid Change Interrupts eIF4E–VPg Interaction in Planta
The yeast two-hybrid assay suggested that the G107R amino acid change in eIF4E-pvr1 alone is sufficient to abolish the capacity of eIF4E to bind VPg. To investigate further the effect of the G107R change on the interaction with VPg, a protein–protein interaction assay in planta was performed using an Agrobacterium tumefaciens–mediated transient expression assay and bimolecular fluorescence complementation (BiFC) assay. The N-terminal half of yellow fluorescence protein (YFP) was fused with the eIF4E construct (YN:eIF4E) and the C-terminal half of YFP was fused with the VPg construct [YC:VPg (TEV-HAT)] (Figure 3A). Yellow fluorescent signal could be detected only when eIF4E and VPg interact or are located closely enough to allow the formation of intact YFP. As shown in Figure 3B, YN:eIF4E-Pvr1+, YN:eIF4E-pvr1, or YN:eIF4E-G107R without YC:VPg (TEV-HAT) were used as a negative control. Coexpression of YN:eIF4E-Pvr1+ and YC:VPg (TEV-HAT) showed a strong yellow fluorescence signal, while the coexpression of YN:eIF4E-pvr1 and YC:VPg (TEV-HAT) together was indistinguishable from the negative control. Coexpression of YN:eIF4E-G107R and YC:VPg (TEV-HAT) also did not show yellow fluorescence signal. The reciprocal combination, YC:eIF4E and YN:VPg (TEV-HAT), displayed similar results (data not shown). This is consistent with the yeast two-hybrid assay and suggests that the G107R amino acid change is also responsible for abolishing VPg binding ability in planta.
Figure 3.
Loss of VPg Binding Ability Caused by the G107R Change in eIF4E Protein Was Confirmed in Planta via Agrobacterium-Mediated Transient Expression Assay in N. benthamiana.
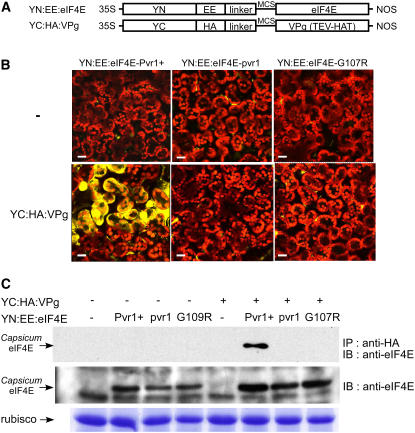
(A) Diagram showing the constructs used in protein–protein interaction assays in planta. The N-terminal half of YFP was fused with eIF4E constructs (YN:eIF4E), and the C-terminal half of YFP was fused with the VPg construct [YC:VPg (TEV-HAT)].
(B) Confocal microscopy images from BiFC assay using N. benthamiana mesophyll cells. eIF4E proteins fused with YN (YN:EE:eIF4E-Pvr1+, YN:EE:eIF4E-pvr1, and YN:EE:eIF4E-G107R) were transiently expressed in N. bethamiana leaf tissue with and without VPg protein fused with YC [YN:HA:VPg (TEV-HAT)]. Yellow fluorescent signal generated by the protein–protein interaction was detected 60 h after infiltration. Chloroplast autofluorescence is shown in red. Bars = 10 μm.
(C) Immunoblot image from coimmunoprecipitation assays. Total protein extracts were pulled down with anti-HA agarose beads and immunoblotted with antibody for Capsicum-eIF4E. Agrobacterium-mediated transient expression of eIF4E protein in N. benthamiana plants was evaluated with the antibody for Capsicum-eIF4E.
Immunoprecipitation using HA tag and Glu-Glu (EE) tag, which were fused downstream of YC and YN, respectively (Figure 3A), was performed to confirm the results of the BiFC assay (Figure 3C). Total protein extracts were immunoprecipitated with anti-HA agarose beads and immunoblotted with an antibody for Capsicum-eIF4E. A strong interaction between YN:EE:eIF4E-Pvr1+ and YC:HA:VPg was detected, whereas YN:EE:eIF4E-pvr1 and YN:EE:eIF4E-G107R were not pulled down with YC:HA:VPg, consistent with the results from the BiFC assay.
Agrobacterium-mediated transient expression of eIF4E protein in Nicotiana benthamiana plants was evaluated with an antibody for Capsicum-eIF4E (Figure 3C). Similar protein accumulation levels were detected for all tissues containing YN:EE:eIF4E-Pvr1+, YN:EE:eIF4E-pvr1, or YN:EE:eIF4E-G107R. Slightly increased protein levels were detected when YN:EE:eIF4E was coexpressed with YC:HA:VPg. Similarly, accumulation of YC:HA:VPg was elevated by coexpression with YN:EE:eIF4E. Interestingly, when a physical interaction occurs between eIF4E and VPg, YC:HA:VPg accumulated to even higher levels (data not shown). VPg interaction activity in planta for eIF4E proteins encoded by other resistance alleles at the pvr1 locus, pvr11 and pvr12, are shown in Supplemental Figure 1 online.
The G107R Substitution Abolishes Cap Binding Ability of eIF4E in Vitro
In a eukaryotic cell, eIF4E recognizes and interacts with the cap structure of cellular mRNA to recruit it into the translation initiation complex. Previously, a cap binding assay using recombinant Capsicum-eIF4E proteins produced in Escherichia coli demonstrated that eIF4E-pvr1 does not show cap binding activity (Kang et al., 2005a). By contrast, eIF4E-pvr11 and eIF4E-pvr12 maintained cap binding ability while resulting in host resistance (Kang et al., 2005a). The cap binding assay using E. coli–produced recombinant protein is a standard method to assess the cap binding ability of a protein; however, it is possible that the recombinant protein failed to retain the characteristics of the native protein produced by host cells. To determine the effect of the variations encoded by resistance alleles on cap binding ability with better resolution and to confirm these results, a cap binding assay using pepper leaf extracts was performed in this study (Figure 4A).
Figure 4.
The Cap Binding Ability of Capsicum eIF4E Proteins Containing Single Amino Acid Changes Was Examined via an in Vitro Cap Binding Assay.
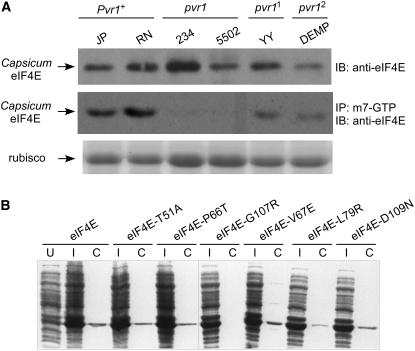
(A) Cap binding assay using protein extracts from six different pepper plants. Immunoblot assay was performed using an antibody for Capsicum-eIF4E before (top panel) and after cap binding assay (middle panel) using the m7-GTP-Sepharose affinity column. The amount of the loaded protein extract was shown by Coomassie blue staining (bottom panel).
(B) Cap binding assay using recombinant eIF4E proteins expressed and extracted from E. coli (DE3). Total cell lysates of E. coli before (U) and after (I) induction by the addition of 20 μM isopropylthio-β-galactoside and proteins showing cap binding activity that are eluted from m7-GTP-Sepharose affinity chromatography (C) were loaded on 12% SDS-PAGE gel and stained with Coomassie blue. The recombinant protein was induced in all E. coli cells containing the eIF4E gene with each substitution. The soluble fraction of bacterial cell lysates containing recombinant eIF4E protein was applied to the m7-GTP-Sepharose column. After washing steps, the protein was eluted from the column using m7-GTP. The eIF4E proteins containing T51A, P66T, V67E, L79R, or D109N were bound to m7-GTP-Sepharose. eIF4E-G107R did not show cap binding activity. U, uninduced; I, induced; C, recombinant eIF4E proteins after m7-GTP-Sepharose affinity chromatography.
In this study, eIF4E-Pvr1+ encoded by two different pepper genotypes, C. annuum JP and RN, appeared to bind strongly to the m7-GTP sephadex columns. By contrast, eIF4E-pvr1 encoded by C. annuum 5502 and Capsicum chinense 234 displayed much reduced affinity for the m7-GTP sephadex columns, consistent with previously published cap binding assays (Kang et al., 2005a). However, both eIF4E-pvr11 and eIF4E-pvr12 also showed a weakened interaction with the cap analog when compared with eIF4E-Pvr1+. The results from this assay support the idea that eIF4E-pvr1, eIF4E-pvr11, and eIF4E-pvr12 all show reduced or compromised cap binding ability relative to eIF4E-Pvr1+, although eIF4E-pvr11 and eIF4E-pvr12 appear to maintain greater cap binding ability than eIF4E-pvr1.
To identify the single amino acid substitutions responsible for the observed disruption of cap binding activity in Capsicum-eIF4E encoded by the recessive resistance alleles at the pvr1 locus, a cap binding assay was performed with eIF4E-T51A, eIF4E-P66T, eIF4E-G107R, eIF4E-V67E, eIF4E-L79R, or eIF4E-D109N (Figure 4B). Recombinant proteins from each construct containing a single substitution were expressed and extracted from E. coli BL21(DE2)pLysS and assayed for binding activity using m7-GTP cap analog columns. The immunoblot shown in Figure 4B shows that all of the eIF4E proteins were reliably expressed after induction using 20 μM isopropylthio-β-galactoside at 20°C and that all constructs showed strong cap binding activity except for eIF4E-G107R. These results indicate that the single amino acid change G107R observed in eIF4E-pvr1 and shown to interrupt the eIF4E-VPg interaction is also responsible for the loss of cap binding ability of eIF4E in vitro. Additionally, L79R, present in both eIF4E-pvr11 and eIF4E-pvr12, negatively affected cap binding ability, although the effect was less pronounced than the complete disruption observed with eIF4E-G107R.
Predicted Protein Models for Mutated eIF4E
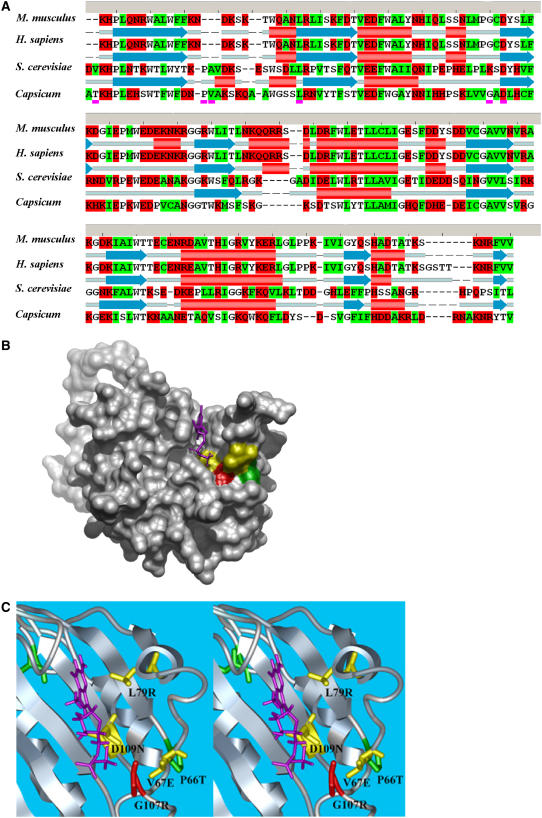
Three dimensional models of the Capsicum-eIF4E protein and the approximate location of the amino acid substitutions were predicted in a previous study based on the murine eIF4E crystal structure (Kang et al., 2005a). This analysis indicated that most of the amino acid substitutions encoded by recessive resistance alleles pvr1, pvr11, and pvr12 are located in the binding pocket of eIF4E. In addition, the amino acid changes are located in close proximity to highly conserved residues that are known to be involved in binding cap or eIF4G. To provide a better understanding of the role of these natural variations at the amino acid level with respect to structural properties of the protein, pairwise structural alignments of the available crystallographic structures of eIF4E from mouse (Protein Data Bank [PDB] code: 1EJ1, monomer A; 190 residues), human (PDB code: 1IPB, monomer A; 217 residues), and yeast (PDB code: 1AP8; 213 residues) were obtained using the Combinatorial Extension method (Shindyalov and Bourne, 1998) and used to optimize the alignment between these protein sequences and that of Capsicum-eIF4E (Figure 5A). The structures of human and mouse eIF4E are quite similar. Optimal superposition of the structures was obtained by aligning 182 residues with no gaps, leading to a root mean square deviation (rmsd) of 1.0 Å for the Cα atoms and a sequence identity of 99.5%. The optimal superposition of the structures of yeast and mouse eIF4E was obtained by aligning 159 residues (with 11 gaps), resulting in an rmsd of 3.3 Å for the Cα atoms and 25.8% sequence identity. Similarly, the optimal superposition of the structures of yeast and mouse eIF4E was obtained by aligning 173 residues (with 12 gaps), resulting in a final rmsd of 3.7 Å for the Cα atoms and 30.1% of sequence identity.
Figure 5.
A 3D Structural Model of Capsicum-eIF4E Protein Showing the Location of Natural Variations in Recessive Virus Resistance Alleles.
(A) Structural alignment of eIF4E proteins from mouse, human, and yeast based on the experimental structures available from PDB (PDB codes 1EJ1, 1IPB, and 1AP8, respectively). The amino acid residues in the sequence are represented by single letters with the background color used to describe the hydrophobic/hydrophilic properties of each particular amino acid. The following convention was used: red = hydrophilic; green = hydrophobic; white = neutral. The assignment of the residues to the secondary structure elements is shown below the sequence: blue arrows are used to indicated β-strands, red rectangles indicate α-helices, and thick gray lines indicate the loop or disordered regions of the chain. The bottom line shows the alignment of the Capsicum-eIF4E sequence encoded by a susceptible allele, Pvr1+, to the experimental structures that were used to generate the 3D models. The mutated residues in the sequence are indicated with a magenta mark below the corresponding letter.
(B) View of the molecular surface area of a model of Capsicum-eIF4E with the amino acid substitutions colored as follows: T51A and P66T in green; G107R in red; V67E, L79R, and D109N in yellow; and 7-methyl-GDP in magenta.
(C) The image zoomed into the hot area of the model (stereoview). The protein is represented using a ribbon model with mutated residues shown using a stick representation. The color codes used are the same as in (B).
The alignment shown in Figure 5A was used to generate all-atom models for the Capsicum-eIF4E protein using the program MODELLER (Sali, 1995; Sali et al., 1995; Sanchez and Sali, 2000). The sequence identity between the final models of Capsicum-eIF4E and the human, mouse, and yeast proteins are 41.2, 35.3, and 35.8%, respectively. Figure 5B shows the molecular surface area of a three-dimensional (3D) model of Capsicum-eIF4E where the amino acid substitutions T51A, P66T, G107R, V67E, L79R, and D109N have been mapped onto that surface. To highlight the location of the binding pocket region, a 7-methyl-GDP molecule (in magenta) is also displayed. In all three resistance alleles, substituted residues occurred within the protein binding pocket with the exception of T51A. Based on this analysis, it seems plausible that the substitutions P66T, G107R, V67E, L79R, and D109N perturb the binding site region, thus affecting cap binding. However, the effects of each substitution on the eIF4E–VPg interaction and cap binding ability appear to be quite different.
The position of several critical substitutions, G107R in eIF4E-pvr1 and V67E and L79R in both eIF4E-pvr11 and eIF4E-pvr12, on the protein binding pocket of eIF4E protein and the positional relationship among these substitutions were estimated using the high-resolution 3D structural model (Figure 5C). The G107R transition in eIF4E-pvr1 appears to be entirely responsible for the loss of in vitro cap binding ability in addition to the interruption of the eIF4E–VPg interaction. To explain how this single amino acid substitution could have such a dramatic effect, we first considered the nature of the substitution. Electrostatic calculations performed with the program GRASP (Nicholls et al., 1991) on a 3D model of Capsicum-eIF4E showed that the region of the molecular surface defining the cap binding slot in Capsicum-eIF4E is covered in its center by low values of electrostatic potential, while the surrounding walls are very positive (data not shown). By changing G107 into Arg, an additional positive charge and a rather large side chain are introduced that may produce strong electrostatic repulsion and steric hindrance with adjacent positively charged residues.
The reduced effects of the V67E and L79R changes found in eIF4E-pvr11 and eIF4E-pvr12 observed for both the VPg–eIF4E interaction and the cap binding relative to the G107R change can be explained as follows. As shown in Figure 5C, residues V67 and L79 lie at both ends of a loop region that lines the binding pocket. This loop contains a key residue, W75, equivalent to murine W56, which was known to be essential for cap binding (Ueda et al., 1991; Marcotrigiano et al., 1997). It appears that the structure can tolerate the presence of a single substitution, either V67E or L79R, because eIF4E-V67E binds to 7-methyl-cap at a similar level to eIF4E-Pvr1+, and eIF4E-L79R can still bind the cap with reduced efficiency. The coexistence of these two substitutions is likely to disrupt the conformation of the loop containing W75 and alter the binding properties of the site. This hypothesis is further supported by the detection of the additive effect of these two changes on the interruption of the eIF4E–VPg interaction.
Transgenic Tomatoes Overexpressing eIF4E-G107R or eIF4E-pvr1+pvr12 Gained Resistance against Several TEV Strains
Ectopic overexpression of Capsicum-eIF4E-pvr1 in S. lycopersicum resulted in gain of viral resistance (Kang et al., 2007). Overexpression of eIF4E-pvr1 in susceptible tomato plants switches the phenotypic outcome from susceptibility to resistance against several potyviruses (Kang et al., 2007). To test the hypothesis that the G107R change in eIF4E-pvr1, which abolishes the physical interaction between eIF4E and VPg in planta, is responsible for gain of resistance, transgenic tomato plants overexpressing eIF4E-G107R were generated and evaluated. Additionally, transgenic plants overexpressing eIF4E-pvr1+pvr12 containing all six substitutions existing in the pvr1 and pvr12 alleles, T55A, P66T, V67E, L79R, G107R, and D109N, were tested for enhancement of resistance and broadened spectrum of resistance.
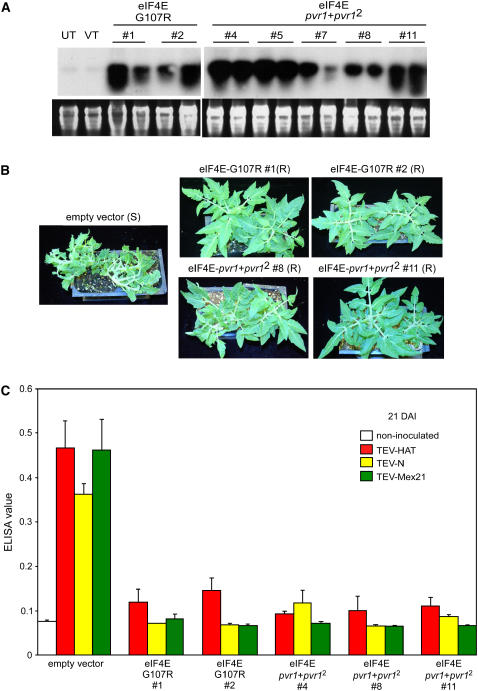
The full-length open reading frames of eIF4E-G107R and eIF4E-pvr1+pvr12 were cloned into the plant transformation vector pBI121 in sense orientation relative to the 35S promoter. Plasmids containing engineered eIF4E constructs were introduced into the experimental tomato variety Micro-Tom via Agrobacterium-mediated transformation (Meissner et al., 1997). Successful transformation was primarily detected by expression of kanamycin resistance. The existence of the transgene was confirmed via PCR screening using two primer sets: one based on the neomycin phosphotransferase II (NPTII) gene and another based on the 35S promoter/eIF4E sequence (data not shown). Progenies derived from self-fertilization of the primary transformants (T0 plants) that segregated in a ratio of 3:1 for resistance/sensitivity to kanamycin, implying that the transgene was integrated at a single locus, were selected for further assays. All T0 plants that contained eIF4E-G107R or eIF4E-pvr1+pvr12 in a single copy appeared normal by visual observation. T1 transgenic progenies obtained by self-pollination from T0 plants were again screened for kanamycin resistance and PCR amplifications. The eIF4E transcript accumulation was assessed in T1 plants by RNA gel blot analysis to provide a relative estimation of the transcription level of the transgene (Figure 6A). All transgenic plants showed eIF4E expression greater than that observed for endogenous eIF4E RNA accumulation. We were not able to detect significantly increased total eIF4E protein in transgenic plants relative to the nontransformed or empty vector–transformed plants via an immunoblot assay using Capsicum-eIF4E antibody. This observation suggests that Capsicum-eIF4E antibody was not capable of distinguishing Capsicum-eIF4E from endogenous Solanum-eIF4E. It is also possible, however, that the levels of eIF4E protein in the transgenic plants are regulated posttranscriptionally. eIF4E is known to be a crucial rate limiting factor for the translation initiation process (Sonenberg and Gingras, 1998); therefore, overexpression may have detrimental effects for the transgenic plants. In fact, transgenic plants containing more than one ectopic copy tended to display reduced growth, although further studies need to be performed to better understand this observation.
Figure 6.
TEV-Screening Results from S. lycopersicum MicroTom T1 Plants Expressing eIF4E-G107R or eIF4E-pvr1+pvr12 for TEV-HAT, TEV-N, and TEV-Mex21.
(A) Expression of eIF4E via RNA gel blot analysis in T1 plants. Total RNA was isolated from each plant and analyzed by RNA gel blot hybridization using pepper eIF4E cDNA labeled with 32P-dCTP as a probe. Ethidium bromide–stained rRNA served as a loading control.
(B) Image of T1 plants containing empty vector, eIF4E-G107R, or eIF4E-pvr1+pvr12 after TEV-N inoculation. Representative phenotypic differences between resistant and susceptible plants are shown. Images were taken at 30 DAI.
(C) ELISA results for T1 plants containing empty vector, eIF4E-G107R, or eIF4E-pvr1+pvr12 after inoculating with TEV-HAT, TEV-N, and TEV-Mex21. Accumulation of TEV coat protein determined by ELISA of tissue sampled from upper uninoculated leaves at 21 DAI. Uninoculated MicroTom plants served as negative controls, and empty vector–containing transgenic plants served as positive controls for viral infection.
Untransformed plants and T1 plants containing eIF4E-G107R or eIF4E-pvr1+pvr12 were used for screening with TEV-HAT, TEV-N, and TEV-Mex21 (Figures 6B and 6C). Three leaves per plant were inoculated with each virus at the four to six leaf stage, and four individual plants representing each genotype were tested. Plants were observed daily for symptom development for more than a month after inoculation. Typical systemic TEV symptoms (terminal leaflet cupping, stunting, and petioles bent downward) developed in all positive control plants, including untransformed plants and T2 plants containing an empty vector, ∼10 d after inoculation (DAI). By contrast, T1 plants carrying eIF4E-G107R or eIF4E-pvr1+pvr12 showed no visible symptoms (Figure 6B). A month after inoculation, T1 plants carrying eIF4E-G107R or eIF4E-pvr1+pvr12 were indistinguishable from their corresponding uninoculated controls. The accumulation of the viral coat protein was assessed using indirect ELISA in uninoculated tissue to detect systemic infection of TEV in the transgenic plants (Figure 6C). The results from ELISA clearly confirmed that T1 plants carrying eIF4E-G107R or eIF4E-pvr1+pvr12 gained complete resistance against all three TEV strains tested: TEV-HAT, TEV-N, and TEV-Mex21. It is interesting to note that while eIF4E-pvr1 transgenic plants showed susceptibility against TEV-Mex21, in accordance with the natural resistance spectrum of the pvr1 allele in pepper (Kang et al., 2007), eIF4E-G107R gained resistance to TEV-Mex21. This suggests that the other two amino acid changes in the engineered resistance allele relative to the wild type, T51A and P66T, actually compromise the potyviral resistance gained by G107R. Transgenic tomatoes containing only T51A and P66T were generated and screened with TEV-HAT, TEV-N, and TEV-Mex21. These transgenic tomatoes remained susceptible to all isolates considered (data not shown). Additionally, transgenic tomatoes carrying eIF4E-pvr11 containing both V67E and L79R displayed delayed susceptibility (see Supplemental Figure 2 online), which is consistent with the observation from pepper genotypes homozygous for the pvr11 allele (Kang et al., 2005a). By contrast, transgenic tomatoes containing the amino acid substitutions from both pvr1 and pvr12 (eIF4E-pvr1+pvr12) resulted in an increased resistance spectrum to include all resistances conferred by either allele alone.
DISCUSSION
The Naturally Occurring Amino Acid Substitution G107R in the pvr1 Allele Appears to Be Responsible for Conformational Changes Leading to Modification of the Cap Binding Pocket
Several lines of evidence indicate that the Gly-to-Arg change at position 107 (G107R) in the Capsicum-eIF4E protein encoded by the pvr1 allele is a critical substitution causing the loss of interaction with TEV-VPg and alone is sufficient for the gain of resistance against potyvirus infection. This observation that the change from a Gly to an Arg appears to be responsible for the disruption of the cap binding pocket has several possible explanations. First, a neutral Gly residue is replaced with a positively charged residue with a relatively large side chain. Amino acid 107 is adjacent to Arg-171, an amino acid that interacts directly with the negative charge of the cap phosphate group and is known to be important for cap binding (Marcotrigiano et al., 1997). The addition of another positive charge in this region is likely to produce repulsion not present in the wild-type protein, causing the location of this important residue to shift. Additionally, this region of the cap binding pocket is rather constrained and may be unable to accommodate the presence of a large residue, such as Arg. The length of the Arg side chain is 50% greater than the distance between the predicted arrangement of G107 and the bound cap; thus, the binding region may be inaccessible due to steric hindrance. In summary, it is plausible that the G107R change in the eIF4E protein could cause a strong electrostatic repulsion with adjacent positively charged residues and/or steric hindrance that interrupts the ability of the protein to bind both cap and VPg.
Further analysis of the nuclear magnetic resonance (NMR) structure of yeast, human, and mouse eIF4E provides better indication as to why the G107R change in Capsicum can be so disruptive. The residue G88, both in mouse and human eIF4E, is the equivalent of G107 in Capsicum-eIF4E. In the available x-ray structures, this G88 residue has been forced into a conformation (φ ∼ +105, ψ ∼ −10) in the Ramachandran plot (Ramachandran and Sasisekharan, 1968) that is rarely occupied by any other amino acid residue. However, based on the NMR structure of yeast eIF4E in complex with 7-methyl-GDP (PDB code: 1AP8), the equivalent position to G107 in Capsicum-eIF4E is occupied by a Lys residue (K90) that is oriented toward the region of the active site where one of the phosphate groups of the 7-methyl-GDP is docked. The K90 residue is observed for eIF4E proteins in Saccharomyces cereveviae and Schizosaccharomyces pombe, implying that an additional positive charge in that particular position in eIF4E can be accommodated in yeast eIF4E. In the NMR structure of yeast eIF4E, where Gly is substituted by Lys (K90), the conformation of residue K90 (φ = +46, ψ = +25) has shifted toward the αL region of the Ramachandran plot from the highly unfavorable region occupied by G88 in mouse and human eIF4E. However, the stress in the loop region seems to be distributed among a few neighboring residues, including L89 (with φ = +36, ψ = −158) and P88 (with φ = −78, ψ = +42), that have been forced into energetically less-favorable regions of the conformational map (Karplus, 1996) and may act to relieve the stress on the loop caused by K90. In the case of the G107R substitution in eIF4E-pvr1, there is no such residue to perform a similar role. This comparative analysis supports the idea that the G107R substitution alone in Capsicum-eIF4E introduces an additional stress that neighboring residues cannot easily accommodate, causing conformational changes that lead to a substantial modification in the cap binding pocket.
Despite these significant consequences of the G107R change, the impact appears to be localized to the VPg and cap binding regions. Overall folding of the mutant protein appears to proceed normally because it maintains the ability to bind to eIF4G (Kang et al., 2007). In many instances, the plant factors that play a role in virus infection are also vitally important to the host. Accordingly, evolution of resistance may be constrained to modifications that disrupt some protein interactions while maintaining others. In fact, selective consequences of single amino acid changes in eIF4E have been explored statistically by measuring a nonsynonymous-to-synonymous nucleotide substitution rate ratio (ω) (J.R. Cavatorta and A.E. Savage, unpublished results). The results indicate that most sites of the eIF4E coding sequence are under purifying selection or neutral evolution, as expected due to the pleiotropic function of eIF4E in translation initiation. However, a small subset of amino acid sites at the eIF4E locus, including position 107, show a molecular signature of strong positive selection that is nonrandomly distributed with respect to sites involved in resistance against viruses.
The VPg Binding and Cap Binding Abilities of eIF4E Are Structurally Related
Two conserved Trp residues in the mammalian eIF4E protein are known to be directly responsible for cap binding of eIF4E by enhancing π-π stacking enthalpy and allowing the localization of the cap structure in the cap binding slot of eIF4E (Marcotrigiano et al., 1997, 1999). It has been noted that the eIF4E proteins encoded by the pvr1 allele from pepper and the sbm1 allele from pea, which both contain the G107R substitution, show reduced cap binding (Gao et al., 2004; Kang et al., 2005a). Recent studies have suggested that the mRNA cap structure and potyvirus VPg at least partially share the protein binding pocket structure of the eIF4E protein or its isoform eIF(iso)4E (Michon et al., 2006; Miyoshi et al., 2006).
We wished to examine the relationship between cap binding and VPg binding regions using the natural variations found in the Capsicum-eIF4E resistance alleles. The cap binding ability of eIF4E is disrupted when these two Trp residues are substituted by Lys residues (Marcotrigiano et al., 1997); therefore, we induced two mutations in wild-type Capsicum-eIF4E, W75L and W121L, and assayed for VPg binding ability. Based on the results from both yeast two-hybrid and BiFC assays in planta, we were able to detect that the VPg binding ability was also lost for Capsicum-eIF4E containing W75L and W121L mutations (data not shown). We also have shown that eIF4E-pvr1, eIF4E-pvr11, and eIF4E-pvr12 produced in pepper have impaired in vitro cap binding ability relative to wild-type eIF4E. These results strongly suggest that the VPg binding domain overlaps with the cap binding domain, although this cap binding assay has not been performed in vivo.
Further evidence that the VPg and cap binding domains overlap is provided by the observation that the amino acid change G107R alone was sufficient to disrupt cap binding as well as the interaction with VPg. The effects of V67E and L79R, however, both found in eIF4E-pvr11 and eIF4E-pvr12, are quite different. These substitutions strongly affect VPg binding while decreasing cap binding only slightly or not at all. Therefore, it appears that these amino acids, which are located at either end of a loop structure, may be in a region involved in VPg binding but do not directly associate with the mRNA 5′ cap.
The results from several independent eIF4E-VPg protein interaction assays and the information from a 3D structural model of Capsicum-eIF4E, in addition to this cap binding assay, allow us to better understand the roles of these natural variations in the acquisition of disease resistance and maintenance of host function. The fact that a single amino acid change, G107R, results in loss of both VPg and cap binding supports the contention that these two binding domains overlap in the eIF4E protein. The observation that other substitutions affect one interaction but not the other suggests that this overlap between binding domains is not complete.
The G107R Change Conferring Potyvirus Resistance Has Evolved Independently in Recessive Resistance Genes Encoding eIF4E within Various Plant Families
We have shown that the Gly-to-Arg change at position 107 (G107R) in the Capsicum-eIF4E protein appears to be responsible for the gain of resistance conferred by the pvr1 allele originally from C. chinense. In plants, a number of alleles at the eIF4E locus conferring resistance against multiple viruses in the family Potyviridae and at least one virus in the family Tombusviridae have been discovered recently. These include pvr1 in pepper (Ruffel et al., 2002; Kang et al., 2005a), mo1 in lettuce (Nicaise et al., 2003), sbm1 in pea (Gao et al., 2004), pot-1 in tomato (Ruffel et al., 2005), rym4/5 in barley (Stein et al., 2005), and nsv in melon (Nieto et al., 2006). It is striking to note that the critical amino acid substitution in eIF4E-pvr1, G107R, also exists at the homologous sites in several other recessive resistance genes: mo1 and sbm1 in lettuce and pea, respectively. Additionally, similar substitutions at or near position 107 are observed in various recessive potyvirus resistance genes encoding eIF4E, including pvr12 (D109R) from C. annuum, mo11 (G107H) from lettuce, and pot1 (M106I) from tomato. Thus, it appears that a G107R change or a very similar change is present in recessive resistance genes pvr1, pvr12, sbm1, mo11, and pot1. The fact that similar substitutions on the eIF4E gene exist quite frequently in various host plant species suggests that phytopathogenic viruses have historically placed at least moderate if not strong selective pressures on diverse hosts and that evolution of resistance at this locus has been an important source of host plant resistance.
The defense strategy whereby a modified version of host eIF4E results in resistance against phytopathogenic viruses appears to have evolved multiple times, providing a highly durable protection from viral infection. Although viral invasion could presumably be disrupted at a number of essential steps, resistance via modification of the eIF4E protein appears to have arisen independently in numerous plant families. eIF4E has important pleiotropic functions in cap binding during initiation of protein translation that must be maintained in virus resistant genotypes. Several possibilities may account for the evolution of resistance alleles with compromised cap binding ability. One factor that may contribute to maintenance of resistance is functional redundancy of cap binding proteins, indicated in Arabidopsis mutational studies of eIF(iso)4E (Duprat et al., 2002). Additionally, eIF(iso)4E encoded at the pvr6 locus is also known to be involved in recessive resistance to Pepper veinal mottle virus in pepper (Ruffel et al., 2006), which also suggests functional redundancy of eIF4E and eIF(iso)4E in this case. Functional redundancy may therefore free eIF4E of its pleiotropic role in protein translation and allow for the evolution of resistance.
The eIF4E proteins encoded by recessive resistance alleles pvr1, pvr12, mo11, and pot1 contain several amino acid substitutions in addition to G107R. In all cases, these accompanying substitutions occur proximal to position 107 based on the 3D structure model, which suggests that these substitutions may act to relieve the electrostatic and steric hindrances predicted to be introduced by G107R. The presence of additional substitutions in eIF4E resistance alleles may represent an additional mechanism to minimize the costs associated with resistance. Although these amino acids are always present with G107R, they have not been observed to increase the resistance spectra of the associated alleles.
Transgenic Approaches Using Engineered eIF4E Based on Natural Variation May Define a Widely Applicable Strategy for Developing Durable, Broad-Spectrum Resistance
Recessive resistance genes are widespread in plants, generally highly durable through decades of deployment across continents and now represent a new source of dominant genetic variation for transgenic deployment. In this article, we show that overexpression of an engineered version of a recessive resistance allele encoding eIF4E in transgenic tomato plants results in a highly resistant phenotype showing dominant inheritance. It has been shown previously that ectopic expression of eIF4E containing natural variations derived from a resistant Capsicum plant perturb interactions required for viral susceptibility in a heterologous system without obvious adverse effects to the host (Kang et al., 2007).
The shift to dominant inheritance suggests that resistant versions of eIF4E may interfere with host factors required for viral infection (Kang et al., 2007). It has been shown that naturally occurring resistance alleles retain an eIF4G binding domain (Kang et al., 2007), implying that eIF4G may be effectively sequestered from the virus. It is also plausible that high levels of overexpressed ectopic eIF4E cause downregulation of native eIF4E. The possibility of posttranscriptional and/or posttranslational regulation of the final expression level of a translation initiation factor has been previously suggested in transgenic Arabidopsis lines overexpressing eIF1A that show tolerance to salt stress (Rausell et al., 2003). This explanation raises the possibility of potential pleiotropic effects of overexpressed eIF4E-G107R, which may also impair cap binding ability necessary for the host. In an attempt to address this question, we have closely monitored two self-pollinated generations of transgenic plants and have observed no visible defects in germination, vegetative growth, flowering and fruit development, or other morphological phenotypes. Consistent with this observation is the fact that the naturally existing recessive resistance alleles pvr1, mo1, and sbm1 in pepper, lettuce, and pea, respectively, contain the G107R changes and show no perceived effects of any type. In addition, unpublished analyses from our lab show that the amino acid position 107 in eIF4E is positively selected through evolution (J.R. Cavatorta and A.E. Savage, unpublished results), a molecular signature that is typically observed in traits with positive fitness consequences. Intriguing questions remain, however, about whether mRNA translation efficiency of the host cell is affected when engineered eIF4E with impaired cap binding ability is overexpressed in transgenic plants. We are currently focusing on differentiating native eIF4E and ectopically expressed eIF4E in the transgenic plants and are also interested in understanding possible pleiotropic effects of the transgenic events.
Viral isolates overcoming the predominant resistance genes have been identified, implying that the viruses coevolved to compensate for the interruption in the VPg–eIF4E interaction (Moury et al., 2004; Kang et al., 2005b). The accumulation of existing variations in resistance alleles encoding eIF4E into one eIF4E construct may stimulate broader-spectrum resistance with increased durability. The mechanism by which the eIF4E–VPg interaction is disrupted may differ in a transgenic system relative to naturally occurring recessive resistance. However, it is likely that transgenic plants expressing appropriate eIF4E alleles would also show stability and durability through large-scale deployment in agriculture.
METHODS
Plant and Virus Materials
Solanum lycopersicum MicroTom seeds used for transformation were obtained from Ball Horticultural Company. Capsicum annuum NuMex RNaky (RN), Early Cal Wonder (ECW), Florida VR2 (VR2), Yolo Y (YY), and breeding line 5502 were obtained from Asgrow Seed Co. C. annuum Dempsey (DEMP) was provided by M. Deom (University of Georgia), and C. annuum Jupiter was provided by Syngenta Seeds. Capsicum chinense PI 159234 (234) was obtained from the USDA Southern Regional Plant Introduction Station. TEV-HAT and TEV-NW (nonwilting on Capsicum frutescens Tabasco) cultures were obtained from T. Pirone (University of Kentucky, Lexington, KY). TEV-N and TEV-Mex21 were obtained from J. Murphy (Auburn University, Auburn, AL). Each recessive resistant allele at the pvr1 locus shows a differential resistance spectrum against these TEV strains: the pvr1 allele is resistant to HAT, NW, and N, the pvr11 allele is resistant to none of these TEV strains, and the pvr12 allele is resistance to HAT, NW, and Mex21. All TEV strains were maintained on TMV-resistant Nicotiana tabacum Kentucky 14. Nicotiana benthamiana plants were used for protein–protein interaction assays in planta.
Protein Extraction Preparation and Immunoblotting Using Plant Tissue
Evaluation of protein expression in plant leaf tissue via immunoblot assay has been described previously (Lin et al., 1995; Menke et al., 2005). For immunoblots, proteins were electrotransferred onto Immun-Blot PVDF membrane (Bio-Rad), and eIF4E protein was detected with an antibody for Capsicum-eIF4E and peroxidase-labeled anti-rabbit antibodies using an ECL kit (GE Healthcare).
Site-Directed Mutagenesis and Plasmid Construction
Cloning of Capsicum-eIF4E alleles and VPg has been described previously (Kang et al., 2005a). The plasmid pGEM-T containing Capsicum-eIF4E was used for site-directed mutagenesis using the QuikChange II-E site-directed mutagenesis kit (Stratagene) following the manufacturer's instructions. Primer sets used for site-directed mutagenesis are listed in Supplemental Table 1 online. Primer sets used for subcloning processes are listed in Supplemental Table 2 online.
The plasmid construction for cap binding analysis using pET16b vector system also has been previously described in detail (Kang et al., 2005a). To generate the fusion protein with eIF4E and either the N-terminal half of YFP tagged with Glu-Glu (YN:EE) or the C-terminal half of YFP tagged with influenza hemagglutinin (HA) epitope tag (YC:HA), the pSY vector series (pSY 736, pSY 735, pSY 728, and pSY 738; provided by Nir Ohad, Tel Aviv University, Tel Aviv, Israel) was used (Bracha-Drori et al., 2004). In the case of VPg, an intragenic BamHI recognition site was erased using site-directed mutagenesis. To express these fusion proteins in planta, the pCAMBIA binary vector system was used and the HindIII restriction enzyme site was selected for subcloning. Plant transformation was performed using the pBI121 system as described previously (Kang et al., 2007). The DNA sequences were confirmed for the resulting plasmids after the subcloning process.
Yeast Two-Hybrid Analysis
Yeast two-hybrid analysis was performed as previously described (Kang et al., 2005a). Yeast strains and plasmid vectors were provided by G.B. Martin (Boyce Thompson Institute, Ithaca, NY). A bait plasmid, pEG202, was used for the fusion of VPg from TEV-HAT, TEV-NW, TEV-Mex21, and TEV-N strains; a prey plasmid, pJG4-5, was used to express Capsicum-eIF4Es containing each substitution separately. Both bait plasmid and prey plasmid were transformed into yeast strain EGY48 containing the lacZ reporter plasmid pSH18-34. The equivalent expression of eIF4E protein in yeast cells was confirmed by immunoblot assay following a procedure previously described (Printen and Sprague, 1994).
Agrobacterium-Mediated Transient Protein Expression in N. benthamiana
The incubation, induction, and coinfiltration of Agrobacterium tumefaciens cultures were performed as previously described with minor modifications (Bendahmane et al., 2000). The cultures were infiltrated into the N. benthamiana leaves at 0.25 OD600.
Fluorescence and Confocal Microscopy Imaging
Protein–protein interaction in planta was monitored 40, 60, and 80 h after Agrobacterium coinfiltration using BiFC assay. The signal from YFP, which is generated by the contact of YN and YC when the two target proteins are proximal, was monitored using a Leica TCS SP2 scanning confocal microscope.
Coimmunoprecipitation
Coimmunoprecipitation was performed as described previously (Moffett et al., 2002; Rairdan and Moffett, 2006). Immunoprecipitated samples were collected and separated by 10% SDS-PAGE gel and blotted onto an ImmunoBlot PVDF membrane (Bio-Rad) for immunoblot assay.
Cap Binding Assays
The cap binding assay was performed as previously described (Morino et al., 1996; Kang et al., 2005a). Expression of the recombinant proteins and purification of the proteins by m7GTP-Sepharose 4.B (Amersham Biosciences) were performed as described previously with minor modifications (Friedland et al., 1997). After eIF4E protein was eluted using the extraction buffer containing 100 uM m7GTP, SDS-PAGE and immunoblot analysis were performed using a Capsicum-eIF4E antibody (New England Biolabs) for detection of eIF4E maintaining cap binding ability in vitro.
Tomato Transformation and Transgenic Screening
The vector construction and tomato transformation followed previously described methods (Kang et al., 2007). S. lycopersicum MicroTom cotyledons were transformed and regenerated into whole plants as described earlier (Meissner et al., 1997). At least 10 T0 transgenic individuals were obtained for each construct. Regenerated plants were self-pollinated and T1 seeds collected. For kanamycin resistance testing, plants at the three or four leaf stage were sprayed with 200 μg/mL of kanamycin solution more than three times. Genomic DNA was extracted using a hexadecyltrimethylammonium bromide method, and the presence of the NPTII gene and eIF4E constructs were verified via PCR amplification using the primers described previously (Kang et al., 2007). For RNA gel blot analysis, total RNA was isolated from tomato leaves using the RNeasy plant mini kit (Qiagen) following the manufacturer's instructions. RNA gel blots were performed according to standard methods. Probes were labeled with 32P-dCTP (Amersham Biosciences) using the Prime It II kit (Stratagene).
Virus Screening Procedure
T1 plants confirmed as transgenic tomatoes were evaluated for viral infection using TEV-HAT, TEV-N, TEV-Mex21, and PepMoV. Plants were inoculated at the four to six leaf stage. After light application of carborundum, viral inoculum was applied to the three oldest leaves. Viral inoculum was produced by grinding systemically infected tobacco tissue in 50 mM potassium phosphate buffer, pH 7.5 (1 g of tissue: 20 mL of buffer). Mock-inoculated and uninoculated controls were routinely included. Treatments typically consisted of four plants. Inoculated plants were monitored for appearance of symptoms, and systemic infection was evaluated at 9 and 21 DAI using antigen plate-coating indirect ELISA as previously described (Kang et al., 2005a). Anti-viral immunoglobulins were obtained from Agdia and used according to the manufacturer's instructions.
Protein Modeling
The program MODELLER (Sali and Blundell, 1993; Sali et al., 1995; Sanchez and Sali, 2000) was used to generate the 3D models for the sequence of Capsicum-eIF4E. The input data for the MODELLER program were as follows: (1) a template structure corresponding to an experimentally determined structure retrieved from the PDB (Berman et al., 2000), and (2) a pairwise alignment between the particular Capsicum sequence and that of the template structure. MODELLER minimizes the violations of distance and dihedral-angle restraints derived from the structures used as templates. Models based on three different templates (mouse, human, and yeast proteins) were built to assess the variability of different parts of the structure, particularly parts involving loop regions where substitutions occur. To facilitate this task, a structure alignment of the experimentally determined structures of the mouse, human, and yeast eIF4E proteins was produced using the Combinatorial Extension method (Shindyalov and Bourne, 1998). Final adjustment of the alignment of the Capsicum-eIF4E with mouse, human, and yeast sequences was performed with the help of graphic tools included in the commercial programs ICM (MOLSOFT) and DS-Modeling (Accelrys). The final manual alignment was used in the model generation process.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AY485127, AY485129, AY485130, AY485131, and AAA48909.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Confocal Microscopy Images from Bimolecular Fluorescence Complementation Assay Using Agrobacterium-Mediated Transient Expression Assay in N. benthamiana.
Supplemental Figure 2. Accumulation of TEV Coat Protein Determined by ELISA for Transgenic Tomatoes Containing Empty Vector, eIF4E-pvr1, or eIF4E-pvr11.
Supplementary Material
Acknowledgments
We thank J.D. Frantz, G.B. Martin, J.F. Murphy, N. Ohad, and S. Yalovsky for experimental materials and E.D. Earle, G. Moriarty, M. Kreitinger, M. Falise, M. Mazourek, S. Roof, and G. Rairdan for technical assistance. We thank S.M. Gray, S.R. McCouch, G.B. Martin, P. Moffett, K. Perez, and G.M. Stellari for useful discussions and critical review of this manuscript. This work was supported in part by USDA National Research Initiative Competitive Grant Program Plant Genome Award 94-37300-0333, USDA Initiative for Future Agriculture and Food Systems Award 2001-52100-113347, and National Science Foundation Plant Genome Award 0218166. I.Y. was supported in part by a fellowship from the Kwanjeong Educational Foundation. Part of this work was conducted by using the resources of the Computational Biology Service Unit from Cornell University, which is partially funded by Microsoft.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Molly M. Jahn (mjahn@cals.wisc.edu).
Online version contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Baulcombe, D. (2004). RNA silencing in plants. Nature 431 356–363. [DOI] [PubMed] [Google Scholar]
- Bendahmane, A., Querci, M., Kanyuka, K., and Baulcombe, D.C. (2000). Agrobacterium transient expression system as a tool for the isolation of disease resistance genes: Application to the Rx2 locus in potato. Plant J. 21 73–81. [DOI] [PubMed] [Google Scholar]
- Berman, H.M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T.N., Weissig, H., Shindyalov, I.N., and Bourne, P.E. (2000). The protein data bank. Nucleic Acids Res. 28 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracha-Drori, K., Shichrur, K., Katz, A., Oliva, M., Angelovici, R., Yalovsky, S., and Ohad, N. (2004). Detection of protein-protein interactions in plants using bimolecular fluorescence complementation. Plant J. 40 419–427. [DOI] [PubMed] [Google Scholar]
- Carrington, J.C., Kasschau, K.D., and Johansen, L.K. (2001). Activation and suppression of RNA silencing by plant viruses. Virology 281 1–5. [DOI] [PubMed] [Google Scholar]
- Cuesta, R., Xi, Q., and Schneider, R.J. (2000). Adenovirus-specific translation by displacement of kinase Mnk1 from cap-initiation complex eIF4F. EMBO J. 19 3465–3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangl, J., and Jones, J.D. (1998). Plant-microbe interactions. Affairs of the plant: Colonization, intolerance, exploitation and co-operation in plant-microbe interactions. Curr. Opin. Plant Biol. 1 285–287. [DOI] [PubMed] [Google Scholar]
- Dangl, J.L., and Jones, J.D. (2001). Plant pathogens and integrated defence responses to infection. Nature 411 826–833. [DOI] [PubMed] [Google Scholar]
- Duggal, R., and Hall, T.C. (1995). Interaction of host proteins with the plus-strand promoter of brome mosaic virus RNA-2. Virology 214 638–641. [DOI] [PubMed] [Google Scholar]
- Duprat, A., Caranta, C., Revers, F., Menand, B., Browning, K.S., and Robaglia, C. (2002). The Arabidopsis eukaryotic initiation factor (iso)4E is dispensable for plant growth but required for susceptibility to potyviruses. Plant J. 32 927–934. [DOI] [PubMed] [Google Scholar]
- Friedland, D.E., Shoemaker, M.T., Xie, Y., Wang, Y., Hagedorn, C.H., and Goss, D.J. (1997). Identification of the cap binding domain of human recombinant eukaryotic protein synthesis initiation factor 4E using a photoaffinity analogue. Protein Sci. 6 125–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale, M., Jr., Tan, S.L., and Katze, M.G. (2000). Translational control of viral gene expression in eukaryotes. Microbiol. Mol. Biol. Rev. 64 239–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, Z., Johansen, E., Eyers, S., Thomas, C.L., Noel Ellis, T.H., and Maule, A.J. (2004). The potyvirus recessive resistance gene, sbm1, identifies a novel role for translation initiation factor eIF4E in cell-to-cell trafficking. Plant J. 40 376–385. [DOI] [PubMed] [Google Scholar]
- Garfinkel, M.S., and Katze, M.G. (1992). Translational control by influenza virus. Selective and cap-dependent translation of viral mRNAs in infected cells. J. Biol. Chem. 267 9383–9390. [PubMed] [Google Scholar]
- Grzela, R., Strokovska, L., Andrieu, J.P., Dublet, B., Zagorski, W., and Chroboczek, J. (2006). Potyvirus terminal protein VPg, effector of host eukaryotic initiation factor eIF4E. Biochimie 88 887–896. [DOI] [PubMed] [Google Scholar]
- Gutierrez, C. (2000). DNA replication and cell cycle in plants: Learning from geminiviruses. EMBO J. 19 792–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez, C., Ramirez-Parra, E., Mar Castellano, M., Sanz-Burgos, A.P., Luque, A., and Missich, R. (2004). Geminivirus DNA replication and cell cycle interactions. Vet. Microbiol. 98 111–119. [DOI] [PubMed] [Google Scholar]
- Ishikawa, M., Diez, J., Restrepo-Hartwig, M., and Ahlquist, P. (1997). Yeast mutations in multiple complementation groups inhibit Brome mosaic virus RNA replication and transcription and perturb regulated expression of the viral polymerase-like gene. Proc. Natl. Acad. Sci. USA 94 13810–13815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janda, M., and Ahlquist, P. (1993). RNA-dependent replication, transcription, and persistence of brome mosaic virus RNA replicons in S. cerevisiae. Cell 72 961–970. [DOI] [PubMed] [Google Scholar]
- Kang, B.-C., Yeam, I., Frantz, J.D., Murphy, J.F., and Jahn, M.M. (2005. a). The pvr1 locus in Capsicum encodes a translation initiation factor eIF4E that interacts with Tobacco etch virus VPg. Plant J. 42 392–405. [DOI] [PubMed] [Google Scholar]
- Kang, B.-C., Yeam, I., and Jahn, M.M. (2005. b). Genetics of plant virus resistance. Annu. Rev. Phytopathol. 43 581–621. [DOI] [PubMed] [Google Scholar]
- Kang, B.-C., Yeam, I., Li, H., Perez, K.W., and Jahn, M.M. (2007). Ectopic expression of a recessive resistance gene generates dominant potyvirus resistance in plants. Plant Biotechnol J. 5 526–536. [DOI] [PubMed] [Google Scholar]
- Karplus, P.A. (1996). Experimentally observed conformation-dependent geometry and hidden strain in proteins. Protein Sci. 5 1406–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasschau, K.D., and Carrington, J.C. (1998). A counterdefensive strategy of plant viruses: Suppression of posttranscriptional gene silencing. Cell 95 461–470. [DOI] [PubMed] [Google Scholar]
- Keller, K.E., Johansen, I.E., Martin, R.R., and Hampton, R.O. (1998). Potyvirus genome-linked protein (VPg) determines Pea seed-borne mosaic virus pathotype-specific virulence in Pisum sativum. Mol. Plant Microbe Interact. 11 124–130. [DOI] [PubMed] [Google Scholar]
- Khan, M.A., Miyoshi, H., Ray, S., Natsuaki, T., Suehiro, N., and Goss, D.J. (2006). Interaction of genome-linked protein (VPg) of Turnip mosaic virus with wheat germ translation initiation factors eIFiso4E and eIFiso4F. J. Biol. Chem. 281 28002–28010. [DOI] [PubMed] [Google Scholar]
- Lellis, A.D., Kasschau, K.D., Whitham, S.A., and Carrington, J.C. (2002). Loss-of-susceptibility mutants of Arabidopsis thaliana reveal an essential role for eIF(iso)4E during potyvirus infection. Curr. Biol. 12 1046–1051. [DOI] [PubMed] [Google Scholar]
- Leonard, S., Plante, D., Wittmann, S., Daigneault, N., Fortin, M.G., and Laliberte, J.F. (2000). Complex formation between potyvirus VPg and translation eukaryotic initiation factor 4E correlates with virus infectivity. J. Virol. 74 7730–7737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, C., Ahmad, M., Gordon, D., and Cashmore, A.R. (1995). Expression of an Arabidopsis cryptochrome gene in transgenic tobacco results in hypersensitivity to blue, UV-A, and green light. Proc. Natl. Acad. Sci. USA 92 8423–8427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcotrigiano, J., Gingras, A.C., Sonenberg, N., and Burley, S.K. (1997). Cocrystal structure of the messenger RNA 5′ cap-binding protein (eIF4E) bound to 7-methyl-GDP. Cell 89 951–961. [DOI] [PubMed] [Google Scholar]
- Marcotrigiano, J., Gingras, A.C., Sonenberg, N., and Burley, S.K. (1999). Cap-dependent translation initiation in eukaryotes is regulated by a molecular mimic of eIF4G. Mol. Cell 3 707–716. [DOI] [PubMed] [Google Scholar]
- Mas, A., Alves-Rodrigues, I., Noueiry, A., Ahlquist, P., and Diez, J. (2006). Host deadenylation-dependent mRNA decapping factors are required for a key step in brome mosaic virus RNA replication. J. Virol. 80 246–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews, M.B. (1990). Control of translation in adenovirus-infected cells. Enzyme 44 250–264. [DOI] [PubMed] [Google Scholar]
- Meissner, R., Jacobson, Y., Melamed, S., Levyatuv, S., Shalev, G., Ashri, A., Elkind, Y., and Levy, A. (1997). A new model system for tomato genetics. Plant J. 12 1465–1472. [Google Scholar]
- Menke, F.L., Kang, H.G., Chen, Z., Park, J.M., Kumar, D., and Klessig, D.F. (2005). Tobacco transcription factor WRKY1 is phosphorylated by the MAP kinase SIPK and mediates HR-like cell death in tobacco. Mol. Plant Microbe Interact. 18 1027–1034. [DOI] [PubMed] [Google Scholar]
- Michon, T., Estevez, Y., Walter, J., German-Retana, S., and Le Gall, O. (2006). The potyviral virus genome-linked protein VPg forms a ternary complex with the eukaryotic initiation factors eIF4E and eIF4G and reduces eIF4E affinity for a mRNA cap analogue. FEBS J. 273 1312–1322. [DOI] [PubMed] [Google Scholar]
- Miyoshi, H., Suehiro, N., Tomoo, K., Muto, S., Takahashi, T., Tsukamoto, T., Ohmori, T., and Natsuaki, T. (2006). Binding analyses for the interaction between plant virus genome-linked protein (VPg) and plant translational initiation factors. Biochimie 88 329–340. [DOI] [PubMed] [Google Scholar]
- Moffett, P., Farnham, G., Peart, J., and Baulcombe, D.C. (2002). Interaction between domains of a plant NBS-LRR protein in disease resistance-related cell death. EMBO J. 21 4511–4519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morino, S., Hazama, H., Ozaki, M., Teraoka, Y., Shibata, S., Doi, M., Ueda, H., Ishida, T., and Uesugi, S. (1996). Analysis of the mRNA cap-binding ability of human eukaryotic initiation factor-4E by use of recombinant wild-type and mutant forms. Eur. J. Biochem. 239 597–601. [DOI] [PubMed] [Google Scholar]
- Moury, B., Morel, C., Johansen, E., Guilbaud, L., Souche, S., Ayme, V., Caranta, C., Palloix, A., and Jacquemond, M. (2004). Mutations in Potato virus Y genome-linked protein determine virulence toward recessive resistances in Capsicum annuum and Lycopersicon hirsutum. Mol. Plant Microbe Interact. 17 322–329. [DOI] [PubMed] [Google Scholar]
- Nicaise, V., Gallois, J.L., Chafiai, F., Allen, L.M., Schurdi-Levraud, V., Browning, K.S., Candresse, T., Caranta, C., Le Gall, O., and German-Retana, S. (2007). Coordinated and selective recruitment of eIF4E and eIF4G factors for potyvirus infection in Arabidopsis thaliana. FEBS Lett. 581 1041–1046. [DOI] [PubMed] [Google Scholar]
- Nicaise, V., German-Retana, S., Sanjuan, R., Dubrana, M.P., Mazier, M., Maisonneuve, B., Candresse, T., Caranta, C., and LeGall, O. (2003). The eukaryotic translation initiation factor 4E controls lettuce susceptibility to the Potyvirus Lettuce mosaic virus. Plant Physiol. 132 1272–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls, A., Sharp, K.A., and Honig, B. (1991). Protein folding and association: Insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins 11 281–296. [DOI] [PubMed] [Google Scholar]
- Nieto, C., et al. (2006). An eIF4E allele confers resistance to an uncapped and non-polyadenylated RNA virus in melon. Plant J. 48 452–462. [DOI] [PubMed] [Google Scholar]
- Nimchuk, Z., Eulgem, T., Holt III, B.F., and Dangl, J.L. (2003). Recognition and response in the plant immune system. Annu. Rev. Genet. 37 579–609. [DOI] [PubMed] [Google Scholar]
- Noueiry, A.O., and Ahlquist, P. (2003). Brome mosaic virus RNA replication: Revealing the role of the host in RNA virus replication. Annu. Rev. Phytopathol. 41 77–98. [DOI] [PubMed] [Google Scholar]
- Panavas, T., Serviene, E., Brasher, J., and Nagy, P.D. (2005). Yeast genome-wide screen reveals dissimilar sets of host genes affecting replication of RNA viruses. Proc. Natl. Acad. Sci. USA 102 7326–7331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Printen, J.A., and Sprague, G.F., Jr. (1994). Protein-protein interactions in the yeast pheromone response pathway: Ste5p interacts with all members of the MAP kinase cascade. Genetics 138 609–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provvidenti, R., and Hampton, R.O. (1992). Sources of resistance to viruses in the Potyviridae. Arch. Virol. Suppl. 5 189–211. [DOI] [PubMed] [Google Scholar]
- Rairdan, G.J., and Moffett, P. (2006). Distinct domains in the ARC region of the potato resistance protein Rx mediate LRR binding and inhibition of activation. Plant Cell 18 2082–2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran, G.N., and Sasisekharan, V. (1968). Conformation of polypeptides and proteins. Adv. Protein Chem. 23 283–438. [DOI] [PubMed] [Google Scholar]
- Rausell, A., Kanhonou, R., Yenush, L., Serrano, R., and Ros, R. (2003). The translation initiation factor eIF1A is an important determinant in the tolerance to NaCl stress in yeast and plants. Plant J. 34 257–267. [DOI] [PubMed] [Google Scholar]
- Ray, S., Yumak, H., Domashevskiy, A., Khan, M.A., Gallie, D.R., and Goss, D.J. (2006). Tobacco etch virus mRNA preferentially binds wheat germ eukaryotic initiation factor (eIF) 4G rather than eIFiso4G. J. Biol. Chem. 281 35826–35834. [DOI] [PubMed] [Google Scholar]
- Robaglia, C., and Caranta, C. (2006). Translation initiation factors: A weak link in plant RNA virus infection. Trends Plant Sci. 11 40–45. [DOI] [PubMed] [Google Scholar]
- Rojas, M.R., Hagen, C., Lucas, W.J., and Gilbertson, R.L. (2005). Exploiting chinks in the plant's armor: Evolution and emergence of geminiviruses. Annu. Rev. Phytopathol. 43 361–394. [DOI] [PubMed] [Google Scholar]
- Ruffel, S., Dussault, M.H., Palloix, A., Moury, B., Bendahmane, A., Robaglia, C., and Caranta, C. (2002). A natural recessive resistance gene against Potato virus Y in pepper corresponds to the eukaryotic initiation factor 4E (eIF4E). Plant J. 32 1067–1075. [DOI] [PubMed] [Google Scholar]
- Ruffel, S., Gallois, J.L., Lesage, M.L., and Caranta, C. (2005). The recessive potyvirus resistance gene pot-1 is the tomato orthologue of the pepper pvr2-eIF4E gene. Mol. Genet. Genomics 274 346–353. [DOI] [PubMed] [Google Scholar]
- Ruffel, S., Gallois, J.L., Moury, B., Robaglia, C., Palloix, A., and Caranta, C. (2006). Simultaneous mutations in translation initiation factors eIF4E and eIF(iso)4E are required to prevent Pepper veinal mottle virus infection of pepper. J. Gen. Virol. 87 2089–2098. [DOI] [PubMed] [Google Scholar]
- Sali, A. (1995). Comparative protein modeling by satisfaction of spatial restraints. Mol. Med. Today 1 270–277. [DOI] [PubMed] [Google Scholar]
- Sali, A., and Blundell, T.L. (1993). Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234 779–815. [DOI] [PubMed] [Google Scholar]
- Sali, A., Potterton, L., Yuan, F., van Vlijmen, H., and Karplus, M. (1995). Evaluation of comparative protein modeling by MODELLER. Proteins 23 318–326. [DOI] [PubMed] [Google Scholar]
- Sanchez, R., and Sali, A. (2000). Comparative protein structure modeling. Introduction and practical examples with modeller. Methods Mol. Biol. 143 97–129. [DOI] [PubMed] [Google Scholar]
- Serviene, E., Jiang, Y., Cheng, C.P., Baker, J., and Nagy, P.D. (2006). Screening of the yeast yTHC collection identifies essential host factors affecting tombusvirus RNA recombination. J. Virol. 80 1231–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shindyalov, I.N., and Bourne, P.E. (1998). Protein structure alignment by incremental combinatorial extension (CE) of the optimal path. Protein Eng. 11 739–747. [DOI] [PubMed] [Google Scholar]
- Sonenberg, N. (1987). Regulation of translation by poliovirus. Adv. Virus Res. 33 175–204. [DOI] [PubMed] [Google Scholar]
- Sonenberg, N., and Gingras, A.C. (1998). The mRNA 5′ cap-binding protein eIF4E and control of cell growth. Curr. Opin. Cell Biol. 10 268–275. [DOI] [PubMed] [Google Scholar]
- Sonenberg, N., and Pelletier, J. (1989). Poliovirus translation: A paradigm for a novel initiation mechanism. Bioessays 11 128–132. [DOI] [PubMed] [Google Scholar]
- Soosaar, J.L., Burch-Smith, T.M., and Dinesh-Kumar, S.P. (2005). Mechanisms of plant resistance to viruses. Nat. Rev. Microbiol. 3 789–798. [DOI] [PubMed] [Google Scholar]
- Stein, N., Perovic, D., Kumlehn, J., Pellio, B., Stracke, S., Streng, S., Ordon, F., and Graner, A. (2005). The eukaryotic translation initiation factor 4E confers multiallelic recessive Bymovirus resistance in Hordeum vulgare (L.). Plant J. 42 912–922. [DOI] [PubMed] [Google Scholar]
- Ueda, H., Iyo, H., Doi, M., Inoue, M., Ishida, T., Morioka, H., Tanaka, T., Nishikawa, S., and Uesugi, S. (1991). Combination of Trp and Glu residues for recognition of mRNA cap structure. Analysis of m7G base recognition site of human cap binding protein (IF-4E) by site-directed mutagenesis. FEBS Lett. 280 207–210. [DOI] [PubMed] [Google Scholar]
- Wittmann, S., Chatel, H., Fortin, M.G., and Laliberte, J.F. (1997). Interaction of the viral protein genome linked of turnip mosaic potyvirus with the translational eukaryotic initiation factor (iso) 4E of Arabidopsis thaliana using the yeast two-hybrid system. Virology 234 84–92. [DOI] [PubMed] [Google Scholar]
- Yoshii, M., Nishikiori, M., Tomita, K., Yoshioka, N., Kozuka, R., Naito, S., and Ishikawa, M. (2004). The Arabidopsis cucumovirus multiplication 1 and 2 loci encode translation initiation factors 4E and 4G. J. Virol. 78 6102–6111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.