Abstract
The gene-for-gene mechanism of plant disease resistance involves direct or indirect recognition of pathogen avirulence (Avr) proteins by plant resistance (R) proteins. Flax rust (Melampsora lini) AvrL567 avirulence proteins and the corresponding flax (Linum usitatissimum) L5, L6, and L7 resistance proteins interact directly. We determined the three-dimensional structures of two members of the AvrL567 family, AvrL567-A and AvrL567-D, at 1.4- and 2.3-Å resolution, respectively. The structures of both proteins are very similar and reveal a β-sandwich fold with no close known structural homologs. The polymorphic residues in the AvrL567 family map to the surface of the protein, and polymorphisms in residues associated with recognition differences for the R proteins lead to significant changes in surface chemical properties. Analysis of single amino acid substitutions in AvrL567 proteins confirm the role of individual residues in conferring differences in recognition and suggest that the specificity results from the cumulative effects of multiple amino acid contacts. The structures also provide insights into possible pathogen-associated functions of AvrL567 proteins, with nucleic acid binding activity demonstrated in vitro. Our studies provide some of the first structural information on avirulence proteins that bind directly to the corresponding resistance proteins, allowing an examination of the molecular basis of the interaction with the resistance proteins as a step toward designing new resistance specificities.
INTRODUCTION
Plants have evolved a complex multilayered defense system to fight pathogens (Jones and Takemoto, 2004; Nurnberger et al., 2004; Chisholm et al., 2006). Effector-triggered immunity involves direct or indirect recognition of pathogen effector proteins by plant surveillance proteins, and subsequent activation of defense mechanisms, such as increased ion fluxes, extracellular oxidative burst, transcriptional responses near the infection sites, and the hypersensitive response (HR) (Chisholm et al., 2006). The pairwise association describing the recognition of the pathogen effectors (termed avirulence [Avr] proteins) within a plant cell by the plant surveillance proteins (termed resistance [R] proteins) has been characterized genetically as the gene-for-gene model of plant disease resistance (Flor, 1971).
Although R proteins can impart resistance to a broad range of pathogens, most can be categorized into two main classes based on their domain structure (Dangl and Jones, 2001). The largest group contains nucleotide binding site (NBS) and leucine-rich repeat (LRR) domains and can be subdivided into coiled-coil–NBS-LRR and Toll-interleukin-1 receptor (TIR)-NBS-LRR proteins based on their N-terminal domain (McHale et al., 2006). The latter class of R proteins can impart resistance to bacterial, viral, fungal, and oomycete pathogens. The second major class of R proteins is proteins with extracellular LRRs (Chisholm et al., 2006); this class confers resistance to fungal and bacterial pathogens.
Resistance proteins can function either by interacting directly with the corresponding Avr protein (the receptor-ligand model) or by sensing specific alterations in the plant that result from the action of Avr proteins (the guard hypothesis) (Jones and Takemoto, 2004; Chisholm et al., 2006). Resistance alleles that function by direct recognition are expected to be under diversifying selection; on the other hand, the R alleles that function through indirect recognition need to develop an association with the target of the effector action, which may preclude their diversification. Both direct (Jia et al., 2000; Deslandes et al., 2003; Dodds et al., 2006; Ueda et al., 2006) and indirect (Axtell and Staskawicz, 2003; Mackey et al., 2003; Shao et al., 2003) recognition has been demonstrated in several independent gene-for-gene systems.
Because R proteins trigger a complex response, including the HR, they need to be tightly regulated. In the absence of an Avr protein, they are therefore likely to be kept in an inactive form either through intramolecular interactions (cis-repression) (Moffett et al., 2002; Hwang and Williamson, 2003) or binding of a repressor protein (trans-repression) (Axtell and Staskawicz, 2003; Mackey et al., 2003; Belkhadir et al., 2004). The Avr proteins are predicted to directly or indirectly induce conformational changes leading to activation of the R proteins (Moffett et al., 2002; Howles et al., 2005; Takken et al., 2006; Ueda et al., 2006).
Pathogens are under evolutionary pressure to evolve diverse Avr proteins that do not induce a resistance response but maintain their core function in the pathogen life cycle. Different effectors do not appear to share common structural features and are thought to perform diverse functions in the host cell. Bacterial Avr proteins are usually delivered directly into the plant cytoplasm via a type III secretion system (Lahaye and Bonas, 2001) and may have enzymatic functions (e.g., protease or ubiquitin ligase) or may activate plant transcription (Chisholm et al., 2006). These bacterial Avr proteins are generally recognized by intracellular R proteins of the NBS-LRR class (Luderer and Joosten, 2001). By contrast, the Avr proteins from extracellular fungal pathogens are usually Cys-rich proteins secreted into the plant intercellular space (apoplast) and display functions such as protease inhibition or chitin binding (van den Burg et al., 2003; Rooney et al., 2005; Chisholm et al., 2006). Such Avr proteins are therefore often recognized by membrane-bound R proteins with extracellular LRRs (Luderer and Joosten, 2001). However, the majority of known R proteins that recognize fungal and oomycete pathogens are cytoplasmic NBS-LRR proteins.
Rust fungi cause disease on many important crop plants, such as cereals and soybean (Glycine max). They are obligate biotrophs that grow only on living plant tissue. In flax (Linum usitatissimum), at least 31 rust resistance specificities to different flax rust (Melampsora lini) strains have been identified, and they are distributed among five polymorphic loci, K, L, M, N, and P (Islam and Mayo, 1990). R proteins encoded by the genes at the L, M, N, and P loci are members of the intracellular NBS-LRR class (Lawrence et al., 1995; Anderson et al., 1997; Dodds et al., 2001a, 2001b). The flax L locus contains at least 12 alleles (Ellis et al., 1999). Diversifying selection has been observed in the LRR domains of the L proteins in particular, as has been observed for essentially all polymorphic NBS-LRR proteins characterized to date; the LRR domains are therefore implicated in controlling recognition specificity (Ellis et al., 1999, 2000; Dodds et al., 2000, 2001a; Jia et al., 2000; Luck et al., 2000; Deslandes et al., 2003; Shen et al., 2003; Jones and Takemoto, 2004).
Several Avr gene families have been cloned from flax rust (Dodds et al., 2004; Catanzariti et al., 2006), including one encoded at the AvrL567 locus. Protein products of AvrL567 genes trigger resistance responses by the L5, L6, and L7 proteins when transiently expressed inside plant cells. These biotrophic fungi can therefore deliver proteins into the plant cytoplasm (Dodds et al., 2004). Many fungi and oomycetes invaginate the host cell plasma membrane with a feeding structure called a haustorium. AvrL567 proteins contain signal peptides and are secreted into the extrahaustorial matrix. Because expression of these proteins in plant cytoplasm induces an HR depending on the presence of cytoplasmic L5, L6, or L7 proteins, there may be a specific translocation mechanism that delivers these proteins into the host cell cytoplasm during rust infection. Most of the Avr proteins from haustorium-forming eukaryotic pathogens characterized to date contain N-terminal secretion signals and trigger HR when localized in the host cell (Allen et al., 2004; Armstrong et al., 2005; Rehmany et al., 2005; Catanzariti et al., 2006; Ellis et al., 2006).
To understand the specificity of the resistance response by different AvrL567 proteins and to obtain clues about their presumed pathogenicity-associated functions, we determined the three-dimensional structures of two AvrL567 variants with different recognition specificities, AvrL567-A and AvrL567-D. The two proteins share 92% amino acid sequence identity (they differ at 12 positions; Figure 1A); they are both functional Avr proteins that differ in the specificity of R protein recognition (Dodds et al., 2004, 2006). AvrL567-D is recognized by L6 but not L5, while AvrL567-A is recognized by both L5 and L6 (Dodds et al., 2006). The structures of both proteins are very similar and reveal a novel β-sandwich fold. An intriguing structural similarity with the protein ToxA from the fungus Pyrenophora tritici-repentis (Sarma et al., 2005) and a demonstration of DNA binding ability of AvrL567 family members give clues about their possible pathogenicity-associated functions. The polymorphic residues in the AvrL567 family map to the surface of the protein and provide a framework for understanding the R protein recognition specificity differences. Mutagenesis of specific surface-exposed amino acids identified several residues with important roles in determining recognition specificity.
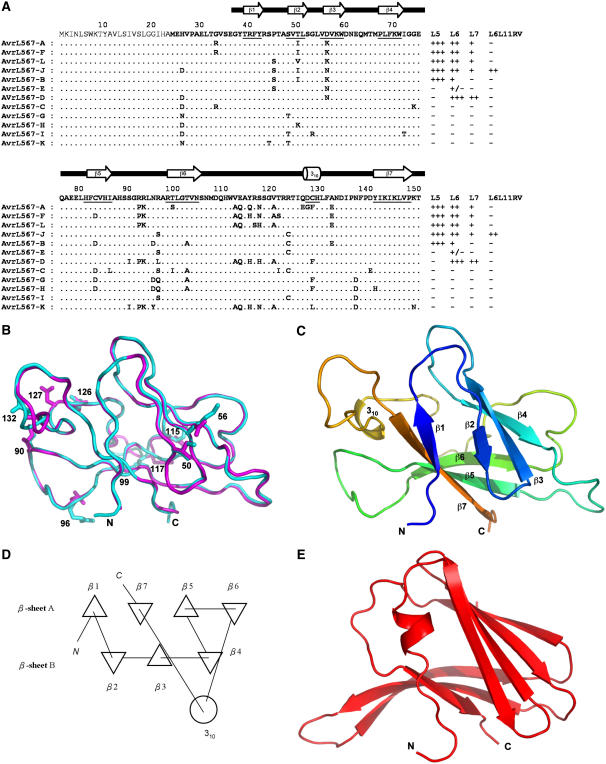
Figure 1.
Structures of AvrL567-A and AvrL567-D Proteins.
(A) Sequence alignment of the known members of the AvrL567 family. Secondary structural elements, based on the structure of AvrL567-A and assigned with the program DSSP (Kabsch and Sander, 1983), are indicated above the sequence and by underlining the sequence. The top row shows the consensus sequence. The signal peptide, shown in thinner font, was not included in the construct used for structure determination. The final columns indicate whether a necrotic response was observed when these proteins were expressed in flax lines containing L5, L6, L7, or the recombinant L6L11RV gene (Dodds et al., 2006). +++ indicates a very strong necrotic response observed within 4 d, ++ indicates necrosis observed within 10 d, + indicates a chlorotic response observed after 10 d, +/− indicates a slight chlorotic response in some but not all assays, and − indicates no response observed.
(B) Superposition of the structures of AvrL567-A (cyan) and AvrL567-D (magenta). The backbones of the proteins are shown in a coil representation, and the side chains of the 10 residues present in the models that differ between the two proteins are shown in a stick representation. This and other structure diagrams were prepared using PyMol (DeLano Scientific).
(C) Ribbon diagram of the structure of AvrL567-A. The β-strands are indicated by arrows and the 310-helix by a coil. The color changes continuously from the N terminus to the C terminus (blue to green and yellow to red).
(D) Schematic diagram of the connectivities of secondary structural elements in AvrL567-A. Triangles represent β-strands, and the circle represents the helix. This was prepared using TOPS (Michalopoulos et al., 2004).
(E) Ribbon diagram of the structure of ToxA (Sarma et al., 2005) after superposition onto AvrL567-A in the orientation shown in (C). The secondary structure elements are shown as in (C).
RESULTS AND DISCUSSION
Structure Determination
All of the 12 AvrL567 gene variants encode predicted proteins of 150 amino acids, including a putative 23–amino acid cleavable signal peptide (Dodds et al., 2004, 2006). The predicted 127–amino acid mature AvrL567-A and AvrL567-D proteins (lacking the signal peptide) were expressed as recombinant proteins in Escherichia coli. Both proteins contained N-terminal hexahistidine and ubiquitin tags, enabling the proteins to be purified using Co2+ affinity chromatography, and the tags were removed using the deubiquitinating enzyme (Dodds et al., 2006). The structure of AvrL567-A was determined by single-wavelength anomalous dispersion techniques using a home x-ray source, taking advantage of a bound Co2+ ion (Guncar et al., 2007). The structure of AvrL567-D was solved by molecular replacement (using the structure of AvrL567-A as a search model). The structures of AvrL567-A and AvrL567-D were refined at 1.4 and 2.3 Å, respectively (Table 1).
Table 1.
Structure Determination
| Diffraction Data Statistics | AvrL567-A | AvrL567-D |
|---|---|---|
| Resolution (Å) | 50.0–1.43 (1.468–1.432)a | 50.0–2.26 (2.316–2.262)a |
| Observations | 337,806 | 102,320 |
| Unique reflections | 28,149 | 7,619 |
| Completeness (%) | 99.1 (93.3) | 99.7 (98.5) |
| Rmerge (%)b | 0.056 (0.238) | 0.113 (0.497) |
| Average I/σ(I) | 33.7 (13.95) | 11.9 (8.35) |
| Refinement statistics | ||
| Resolution (Å) | 42.1–1.43 (1.468–1.432) | 40.26–2.26 (2.316–2.262) |
| Number of reflections | 26,436 | 7,199 |
| Rcrystc | 0.220 (0.387) | 0.195 (0.240) |
| Rfreed | 0.254 (0.384) | 0.257 (0.352) |
| Number of nonhydrogen atoms: | ||
| Protein | 1104 | 913 |
| Solvent | 199 | 103 |
| Mean B-factor (Å2) | 24.6 | 28.4 |
| Coordinate error (Å) | 0.198 | 0.250 |
| Root mean square deviations from ideal values: | ||
| Bond lengths (Å) | 0.015 | 0.019 |
| Bond angles (°) | 1.617 | 1.702 |
| Ramachandran plot: | ||
| Residues in most favored (disallowed) regions (%)e | 88.9 (0) | 87.9 (0) |
Numbers in parenthesis are for the highest resolution shell.
Rmerge = ∑hkl(∑i(|I hkl,i − <I hkl >|))/∑hkl,i <I hkl>, where I hkl,i is the intensity of an individual measurement of the reflection with Miller indices h, k, and l, and <Ihkl> is the mean intensity of that reflection. Calculated for I > −3σ(I).
Rcryst = ∑hkl(‖Fobshkl| − |Fcalchkl‖)/|Fobshkl|, where |Fobshkl| and |Fcalchkl| are the observed and calculated structure factor amplitudes.
Rfree is equivalent to Rcryst but calculated with reflections (5%) omitted from the refinement process.
Calculated with the program PROCHECK (Laskowski et al., 1993).
fCalculated based on the Luzzati plot with the program SFCHECK (Vaguine et al., 1999).
Overall Structure
The structures of AvrL567-A and AvrL567-D are very similar to each other (Figure 1B); after superposition, the root mean square deviation of 113 structurally equivalent Cα atoms is 0.59 Å; therefore, the structure of AvrL567-A will be used for all analyses unless indicated otherwise. The structure is a β-sandwich dominated by two antiparallel β-sheets, sheet A with four strands (β-strands β1, β7, β5, and β6) and sheet B with three strands (β-strands β2, β3, and β4; Figures 1C and 1D) that arrange into an incompletely closed β-barrel. There is also a three-residue 310 helix in the loop connecting β-strands 6 and 7. The N-terminal regions in both AvrL567-A and AvrL567-D (residues 24 to 35 and 24 to 36, respectively) had uninterpretable electron densities, suggesting that these regions are flexible in both structures (the presence of these residues was demonstrated by mass spectrometry analysis on crystals). This is consistent with limited proteolysis results (Dodds et al., 2006), suggesting that the region N-terminal to residue 36 is flexible and accessible to protease digestion in solution. By contrast, the C-terminal region is well ordered with only one C-terminal residue of AvrL567-D exhibiting uninterpretable electron density. The loop comprising residues 118 to 121 has no interpretable electron density in AvrL567-A but is well ordered in AvrL567-D. The structures are consistent with the circular dichroism spectra, which indicated a similar, β-sheet–containing structure for AvrL567-A and AvrL567-D (Dodds et al., 2006). E. coli–expressed AvrL567-C protein also showed a similar circular dichroism spectrum and a protease-accessible N-terminal region (Dodds et al., 2006). Thus, the available structural data and the high degree of sequence similarity among the family members suggest that all 12 members of the AvrL567 family have very similar three-dimensional structures.
Structure-Based Insights into Pathogenicity-Associated Functions of AvrL567 Proteins
The AvrL567 proteins have no significant sequence similarities with any proteins of previously known structure (Dodds et al., 2006). Structure comparisons of AvrL567-A and AvrL567-D with known protein structures (Holm and Sander, 1998; Shindyalov and Bourne, 1998; Laskowski et al., 2005) give the best match with the structure of the protein ToxA from the fungal pathogen P. tritici-repentis that causes tan spot of wheat (Triticum aestivum; Sarma et al., 2005). While the two structures are not closely related (Z-score = 5.3 using the program Dali [Holm and Sander, 1998]; root mean square deviation = 3.1 Å for 83 structurally equivalent Cα atoms), the polypeptide chain topologies are analogous (Figure 1E). The largest structural differences occur in the long connection between the equivalent of AvrL567-A β-strands β6 and β7, which contains a 310 helix in the case of AvrL567-A, while it forms an extra β-strand (inserted into the equivalent of AvrL567-A β-sheet B) in ToxA. There are no similarities with any known structures of avirulence proteins (van den Hooven et al., 2001; van't Slot et al., 2003; Lee et al., 2004; Wulf et al., 2004; Zhu et al., 2004; Janjusevic et al., 2006).
The similarity to ToxA is intriguing because ToxA is a host-selective toxin secreted into the host apoplast by certain races of P. tritici-repentis. Recent work has shown that the apoplastic ToxA protein is internalized into mesophyll cells of sensitive but not insensitive host genotypes, where it induces cell death (Manning and Ciuffetti, 2005). The uptake of this protein is regulated by a single polymorphic host gene, Tsn1, which confers toxin sensitivity, and may encode a receptor allowing endocytosis of the toxin. Mutagenesis experiments with ToxA have implicated an Arg-Gly-Asp (RGD) motif and surrounding sequence in its uptake (Ciuffetti et al., 1997; Manning et al., 2004; Sarma et al., 2005). The structural similarity between ToxA and fibronectin type III domains, classic mammalian RGD-containing sequences, suggests that uptake may be mediated by interactions with plant integrin-like receptors (Sarma et al., 2005). AvrL567 proteins are also predicted to be imported into plant cells during infection, and the structural similarities suggest that there may be some parallels in their uptake mechanism, although the AvrL567 proteins do not contain RGD sequences. The toxicity mechanism of ToxA is not well understood, so it is difficult to draw any functional conclusions regarding AvrL567 from this comparison.
The metaserver ProKnow (Pal and Eisenberg, 2005) identified catalytic activity as the most likely function and GTP binding as another possible function for AvrL567 proteins, based on the structure and the available functional information on related proteins. The ProFunc program (Laskowski et al., 2005), however, did not identify any similarities with enzyme-active site templates. AvrL567-A cocrystallized with a Co2+ ion, derived from the affinity matrix or the crystallization solution. The metal is tetrahedrally coordinated by the side chains of residues His-85 and Cys-83 of one AvrL567-A molecule and His-105 from the neighboring AvrL567-A molecule in the crystal and one imidazole molecule from the solvent. Cys-83 is not strictly conserved in the AvrL567 family and is replaced by Asp in five members of the family. However, it is possible that physiologically relevant metals may bind in this position. We tested the effect of several divalent metals on the behavior of AvrL567-A, AvrL567-C, and AvrL567-D in size-exclusion chromatography (see Methods). An effect was observed with Co2+ (which induced a shift to higher molecular weight, consistent with dimerization, for AvrL567-A but not AvrL567-C and AvrL567-D) and Zn2+ (which induced precipitation of all proteins; data not shown). Because the juxtaposition of the residues 83, 85, and 99 was reminiscent of a Cys-His-Gln catalytic triad in Cys proteases, we tested for protease activity toward a papain substrate, but no activity could be detected (data not shown). Therefore, the significance of metal binding and the existence of any catalytic activity remain to be established.
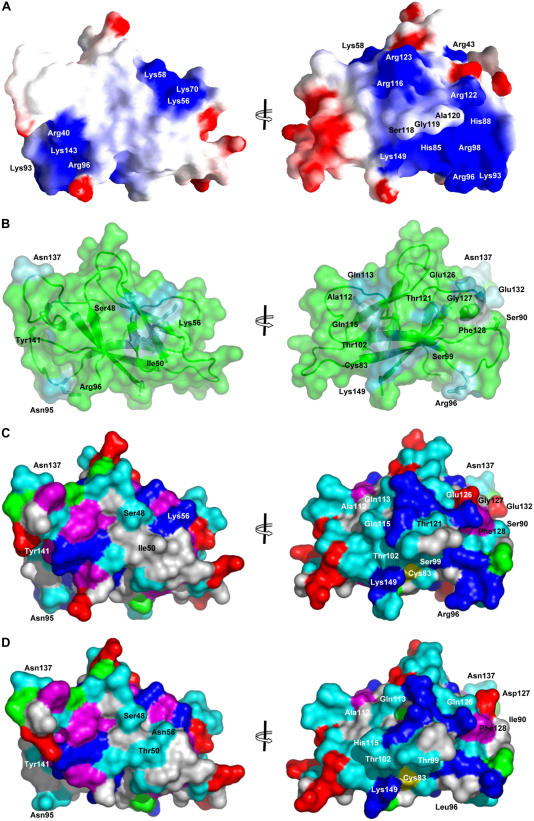
Protein sequence comparisons (Altschul et al., 1990) identified a hypothetical protein from Cryptosporidium hominis and a protein from Cryptosporidium parvum annotated as DNA polymerase ε subunit B (Abrahamsen et al., 2004) as best matches to any protein outside the AvrL567 family, both sharing 25% identity with AvrL567-A. Although the physiological roles of DNA polymerase ε are not fully understood, it is one of three essential replicative polymerases in eukaryotic cells (Shcherbakova et al., 2003). The ProFunc program (Laskowski et al., 2005) identified similarity with a Y-family DNA polymerase (Ling et al., 2001). Inspection of the electrostatic potential mapped to the surface of AvrL567 proteins (Figure 2A) reveals two large positively charged patches interrupted by the protruding loop containing residues 118 to 120. Both patches could potentially serve as DNA binding sites. DNA binding residues predicted by DP-Bind (Hwang et al., 2007) and PreDS (Tsuchiya et al., 2005) are also consistent with these patches. However, AvrL567-C and a number of other variants have much less pronounced positive electrostatic potential on their surface.
Figure 2.
Surface Properties of AvrL567 Variants.
(A) Molecular surface of AvrL567-A color-coded according to electrostatic potential mapped to it. Lys and Arg were assigned a single positive charge, and Glu and Asp were assigned a single negative charge. A uniform dielectric constant of 80 was assumed for the solvent and 2 for the protein interior; the ionic strength was set to zero. Coloring is continuous going from blue (potential +5 kt/e; 1 kt = 0.6 kcal, e is the charge of an electron) through white to red (potential −5 kt/e). This was calculated and drawn with the program GRASP (Nicholls and Honig, 1991). Two views are shown, rotated 180° around the y axis; the left view is the same as in Figure 1B.
(B) The degree of amino acid conservation mapped onto the surface of AvrL567-A, as scored by the program Consurf (Landau et al., 2005). The structure of AvrL567-A is shown in a ribbon representation, and the surface of the protein is shown in a transparent representation. The majority of the protein is colored green, and the nonconserved residues are colored from dark (higher degree of conservation) to light blue (lower degree of conservation). The side chains of selected residues are labeled and shown in stick representation. Two views are shown, rotated 180° around the y axis as in (A).
(C) Surface of AvrL567-A, colored by amino acid properties: hydrophobic, white; aromatic, magenta; polar, cyan; positively charged, blue; negatively charged, red; Cys, yellow; Pro, green. Two views are shown, rotated 180° around the y axis as (A).
(D) Surface of AvrL567-D, shown as in (C).
To test the hypothesis that AvrL567 proteins bind DNA, we performed electrophoretic mobility shift assays using the recently described pentaprobe reagents (Kwan et al., 2003). Pentaprobe comprises six 100-bp double-stranded DNA oligonucleotides that together contain all possible 5-bp DNA sequences and can act as a convenient screen for DNA binding activity in novel proteins. The results show that AvrL567-A and to a lesser extent AvrL567-D (but not AvrL567-C) reduced the mobility of the DNA probe, indicative of DNA binding (see Supplemental Figure 1 online). Binding was also observed to single-stranded DNA and both single- and double-stranded RNA (see Supplemental Figure 1 online). If indeed AvrL567 proteins functioned as DNA or RNA binding proteins, one would expect that all members of the AvrL567 family would share this function, assuming it was important for the pathogen. Furthermore, the lack of strict sequence specificity of binding argues against a function as transcriptional regulator. Thus, the biological significance of the observed nucleic acid binding activity clearly remains to be established.
Structural Basis of the Specificity of Resistance Response
The members of the AvrL567 family display differences in recognition specificity by the corresponding R proteins. Seven variants induce a necrotic response when expressed in flax plants containing corresponding R proteins, with some differences in recognition specificity, and interact with the same corresponding R proteins in yeast two-hybrid assays (Dodds et al., 2006) (Figure 1A). The remaining five AvrL567 variants, derived from virulent rust strains, are not recognized by these R proteins in plants or in yeast. We have suggested previously that the amino acid side chain differences directly affect the R–Avr protein interactions (Dodds et al., 2006). The similarity of the structures of AvrL567-A and AvrL567-D provides additional support to this hypothesis. The knowledge of the structure now allows us to examine the structural basis for the recognition specificities.
The side chains of all but one of the 35 polymorphic positions in the AvrL567 family are highly solvent exposed in the AvrL567-A and -D structures, and substitutions in these residues would not be expected to disrupt protein folding or stability (Figure 2B). The one exception is Ile-86; however, the conservative replacement of this residue by Leu in AvrL567-C is expected to have little effect on structure or stability, which is supported by the structural characterization of AvrL567-C (Dodds et al., 2006). The most likely protein interaction patch predicted by the program ProMate (Neuvirth et al., 2004) involves the loop containing the polymorphic residues Ala-132, Asn-137, and Asp-140 (Figure 1A). A number of polymorphic residues form a prominent narrow patch winding around the structure that includes residues 50 and 56. Based on sequence comparisons, residues 50, 56, 90, and 96 seem to be the most likely polymorphisms among the AvrL567 variants that may explain the differences in recognition by L5 and L6 proteins (Dodds et al., 2006) (Figures 1 and 2). Unique polymorphic residues occur at positions 90 and 96 in AvrL567-D, while the Thr-50 polymorphism is shared with AvrL567-E, which shows a similar specificity (Figure 1A). The five unrecognized variants AvrL567-C, -G, -H, -I, and -K all share an Asp at position 56 that is absent from the other proteins. Polymorphisms in these four residues stand out as leading to significant differences in local surface properties between the AvrL567-A and -D proteins (Figures 2C and 2D) but occur at distant positions. The side chains of residues 50 and 56 are located in β-sheet B (β-strands β2 and β3, respectively), while side chains of residues 90 and 96 are located in the loop connecting β-strands β5 and β6, ∼30 Å away from residues 50 and 56 (Figure 2). A hydrophobic residue is present at position 50 in all proteins that bind L5 but is substituted by a polar residue in all variants that do not. Residue 56, located next to residue 50 on the surface of the protein, is a positively charged or neutral polar residue in all the proteins that bind to L5 or L6 but a negatively charged residue in all the variants that do not. Hydrophobic surface residues at positions 90 and 96 are unique to AvrL567-D and may contribute to its unique specificity. A positively charged residue at position 96 is found in most L5 binding variants but is not strictly required, as demonstrated by Ser substitution in AvrL567-J. In addition to these residues, the substitutions in positions 126 and 127 (unique to AvrL567-A) have a large effect on the local surface properties, although they do not correlate with any specificity differences among the AvrL567 variants.
Identification of Specificity-Determining Residues by Mutational Analysis
To test the hypothesis that amino acid differences at positions 50, 56, 90, and 96 determine differences in recognition specificity, we made targeted substitutions of these amino acids in the AvrL567-A, -C, and -D proteins. Of these three proteins, AvrL567-A is recognized by both L5 and L6, AvrL567-D by L6 only, and AvrL567-C by neither resistance protein (Figure 3A). The mutant AvrL567 proteins were assayed for interactions with L5 and L6 by yeast-two-hybrid analysis and by transient expression in planta (Figures 3B and 3C). Protein immunoblot analysis showed that all of the mutant fusion proteins accumulated to similar levels in yeast (data not shown), indicating that any observed effects on recognition were due to differences in interaction with the corresponding R protein fusions rather than altered stability of the mutant Avr protein fusions.
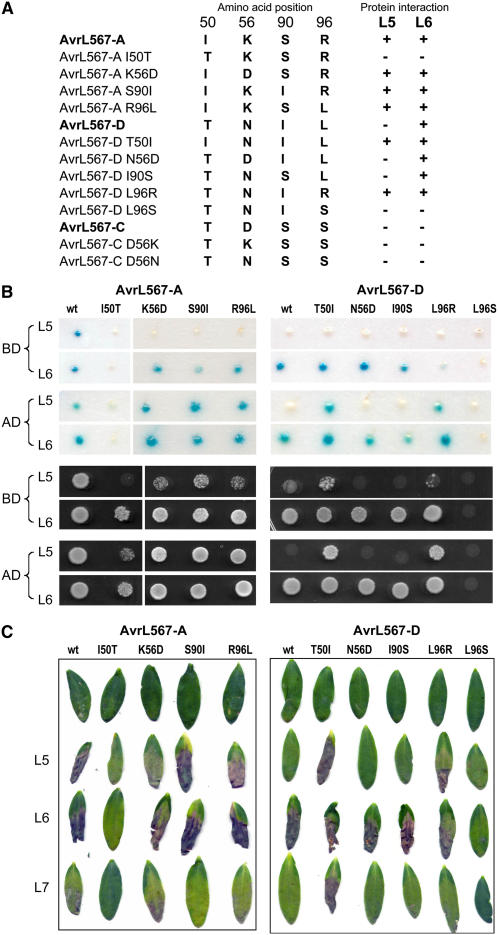
Figure 3.
Mutational Analysis of AvrL567 Interactions with Flax Resistance Proteins.
(A) The amino acid residues present at positions 50, 56, 90, and 96 in the AvrL567-A, -C, and -D and mutant proteins is indicated along with the observed recognition specificity with respect to the L5 and L6 resistance proteins.
(B) Yeast two-hybrid analysis of the interaction among L5, L6, and AvrL567 mutant proteins. Top panels show β-galactosidase activity of yeast strain SFY526 expressing the GAL4 DNA binding domain (BD) or activation domain (AD) fused to the L5 or L6 proteins along with the corresponding GAL4 domain fused to AvrL567-A, -D (wt), or their single amino acid substitution derivatives. Bottom two panels show growth of strain HF7c expressing the same protein fusion constructs on media lacking His.
(C) The AvrL567-A, -D, and single amino acid substitution derivatives were transiently expressed by Agrobacterium infiltration in leaves of flax lines Bison (contains L9) and the L5, L6, and L7 near-isogenic lines. Images were taken 8 d after infiltration.
Reciprocal exchanges of each of the 50, 90, and 96 polymorphisms revealed that two of these positions, 50 and 96, are particularly important for L5 recognition. Substituting either of these residues in AvrL567-D with the corresponding residue from AvrL567-A (T50I or L96R) was sufficient to allow the interaction of this protein with L5 in yeast (Figure 3B). This interaction was detected most strongly when the GAL4-AD was fused to L5 and the GAL4-BD fused to AvrL567 but was weaker when the fusions were reversed. This is consistent with previous indications that the former fusion protein orientation provides a more sensitive measure of this protein interaction (Dodds et al., 2006) and suggests that the L5 interaction may be slightly weaker than the L6 interaction. Nevertheless, expression of these two protein variants in flax led to an L5-dependent HR response (Figure 3C), indicating that the physical interaction detected by yeast two-hybrid analysis correlates with the induction of HR in plant cells. Conversely, the I50T reciprocal substitution in AvrL567-A knocks out recognition by L5 almost completely, both in yeast and in planta, which confirms the importance of this residue in L5 recognition. Both the S90I and R96L substitutions in AvrL567-A have only a slight effect in reducing the L5 interaction in yeast (only observed in the less-sensitive L5-BD/Avr-AD fusions) but no effect in planta, suggesting that the slightly reduced physical interaction between these proteins is still sufficient for effective L5-mediated resistance activation in flax.
Interestingly, most of these changes in AvrL567-A and -D had no effect on L6 recognition, which suggests that the L5 and L6 interactions with these proteins may involve different amino acid contact points. However, the I50T mutation in AvrL567-A substantially decreased the L6 interaction in yeast and also reduced the HR induction in planta. This observation was unexpected because the Thr-50 polymorphism was derived from the AvrL567-D protein, which is recognized by L6; however, the amino acids at the polymorphic positions 50, 56, 90, and 96 in the AvrL567-A I50T mutant closely resemble the corresponding amino acids in AvrL567-C, which similarly does not interact with L5 or L6. Interestingly, the reciprocal substitution (T50I) in AvrL5676-D showed an increased necrotic response when expressed in flax leaves containing L7. This is significant because L7 functions as a weak allele of L6 (i.e., it recognizes all the same AvrL567 variants as L6 but gives a weaker HR response; Dodds et al., 2006), and the L7 protein is identical to L6 except for 11 amino acid changes in the TIR domain. A likely explanation for this observation is that the L6 response to AvrL567-D is likely to be already at or above a saturation point, and any increased binding affinity does not lead to an increase in resistance signaling. However, the weaker response induced by L7 is enhanced by an increased interaction with the Thr-50 mutant. Thus, it appears that the Thr-50 residue in AvrL567-D is actually destabilizing for the L6 interaction but that its effect is masked in this protein by the presence of other stabilizing amino acid contacts.
Although the Arg-Leu polymorphism at position 96 had no apparent effect on L6 recognition, this amino acid position is important for L6 recognition because a Ser substitution at this position in AvrL567-D abolished interaction with L6 in yeast as well as HR induction in flax (Figures 3B and 3C). This polymorphism is present in another AvrL567 variant (J) that is recognized by L6 (Figure 1A), again highlighting that the effect of individual amino acid substitutions on recognition depends on the context of the other polymorphisms present in a particular AvrL567 protein.
We also examined the role of the polymorphism at position 56 that distinguishes the virulence alleles (C, G, H, I, and K). The K56D substitution in AvrL567-A had no effect on recognition by L6 but did have a small effect on L5 interaction in yeast and led to a slightly reduced HR response in L5 plants (Figure 3). The corresponding substitution, N56D, in AvrL567-D had no effect on L6 recognition in yeast or in planta. The reciprocal substitutions in AvrL567-C, D56N and D56K, did not lead to any interaction with L5 or L6 in yeast or in planta (data not shown). Thus, while this polymorphism may have some effect on L5 interaction, it is not the primary cause of the nonrecognition of the AvrL567-C protein or the products of the other virulence alleles of AvrL567. It is likely that other amino acid polymorphisms in these proteins prevent their recognition, with the Thr-50 polymorphsim in AvrL567-C, -G, -I, and -K likely to have a strong effect. AvrL567-C also contains the Ser-96 polymorphism that abolished L6 recognition of AvrL567-D. The presence of both of these polymorphisms in AvrL567-E may also explain its very weak recognition by L6.
In summary, the mutagenesis data suggest that of the four positions tested, the most important residues for determining the specificity are residues 50 and 96. Ile at position 50 is favorable for the interaction with L5, while Thr at this position destabilizes the interaction with L5 and L6. At position 96, Arg favors interaction with L5, while Leu favors interaction with L6. Positions 56 and 90 are shown to play a less significant role, with Asp-56 unexpectedly shown to have only a weak destabilizing effect on L5 and L6 recognition, and Ser-90 and Ile-90 showing only slight preference for L5 and L6, respectively. Overall, these results suggest that the interaction between AvrL567 proteins and their corresponding R proteins involves multiple contact points that have additive effects on the strength of the interaction. The cumulative effect of either stabilizing or destabilizing interactions at each position would determine the overall strength of the interaction. This model can explain how the impact of specific amino acid residues (such as Thr-50 and Ser-96) can depend on the context of the protein, and substitutions at different positions (e.g., Ile-50 or Arg-96 in AvrL567-D) can lead to a similar effect (recognition by the same R protein).
A Structural Model for the AvrL567-A/L6 Interaction
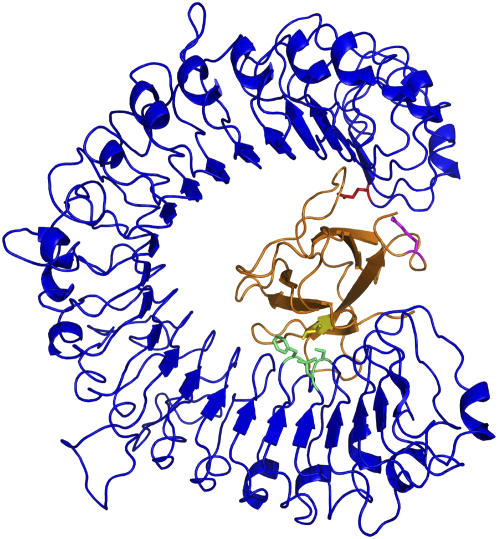
The most likely AvrL protein binding site on L proteins is the LRR domain, based on role of this domain in conferring gene-for-gene specificity as revealed by sequence comparisons and domain swap experiments on L variants and other NBS-LRR proteins (Ellis et al., 1999, 2000; Dodds et al., 2000, 2001a; Jia et al., 2000; Luck et al., 2000; Deslandes et al., 2003; Shen et al., 2003; Jones and Takemoto, 2004). The locations of the amino acid variations on both AvrL567 and L proteins suggest a large surface area involving multiple contacts and therefore likely involves the curved β-sheet of the LRR region of the L proteins (Kobe and Deisenhofer, 1995). In agreement with this idea, selection for amino acid variation is highest in the β-strand/turn motif of LRR units in L and other LRR-containing R proteins (Dodds et al., 2000; Ellis et al., 2000). To test the hypothesis on the location of the binding site, we created comparative models (Fiser and Sali, 2003) of the LRR region of L5 and L6 proteins (see Methods) and docked AvrL567-A and AvrL567-D to this structure using the program Zdock (Chen and Weng, 2002). The models with best scores support the curved β-sheet as the most likely interaction site. The curvature allows for a large interface and facilitates the simultaneous interaction of distant regions of the AvrL567 proteins implicated in the interaction, such as the segments comprising residues 50 to 56 and residues 90 to 96. The example of the model of L5 LRR domain-AvrL567-A complex shown in Figure 4 satisfies the interaction of Ile-50 with a hydrophobic pocket on L5 and of Arg-96 with a negative surface patch on L5. Furthermore, sequence differences between L5 and L6 mapping to these interacting regions (e.g., Glu-1242 in L5 and Leu-1232 in L6) are consistent with the specificities for different AvrL567 variants.
Figure 4.
Model of the Complex between AvrL567-A and the LRR Domain of L5.
An example docked model of the AvrL567-A protein (orange) with the model of the LRR domain of the L5 protein (blue) is shown, highlighting AvrL567-A residues Ile-50 (yellow) and Arg-96 (magenta) and L5 residues Leu-721, Ser-722, Phe-746 (green), and Glu-1242 (red).
Conclusions
Our studies provide structural information on Avr proteins that bind directly to the corresponding NBS-LRR resistance proteins (the largest class of R proteins), allowing an examination of the structural basis of this protein recognition event. The structural and mutagenesis results show that binding between the R and Avr proteins involves multiple contact points that occur across a large surface area of the Avr protein. This analysis supports a structural model in which recognition is mediated through the LRR domain of the R protein, with docking predicting binding of the AvrL567 protein within the predicted curved β-sheet of the LRR domain. Superimposed on this overall structural compatibility between the LRR domain and AvrL567 proteins is the formation of specific amino acid contact points that contribute to stabilizing the protein interaction. The data show that individual amino acid contacts have additive effects in either stabilizing or destabilizing the protein interaction and that it is the summation of these effects that determines the binding affinity of the interaction. There is also a strong correlation between the strength of the R–Avr binding interaction and the strength of the HR response induced in flax (Figure 3) (Dodds et al., 2006). This suggests that activation of the R protein signaling function is a dynamic process that depends on either continuous interaction with the corresponding Avr protein to maintain the R protein in an active signaling conformation or repeated cycles of Avr binding and dissociation associated with multiple discrete signaling events. Either possibility appears consistent with current models of R protein signaling (Rairdan and Moffett, 2006; Tameling et al., 2006).
These results have interesting implications for the coevolution of R and Avr proteins. We previously observed a high level of diversifying selection acting on both the L and AvrL567 genes and suggested that this was due to a coevolutionary arms race (Dodds et al., 2001b, 2006). The observation that single amino acid changes can have quantitative and qualitative effects on both the protein interaction and resistance response allows for a model of stepwise evolution of new R and Avr gene variants. Given the initial shape complementarity between the R and Avr proteins, single amino acid changes in specific LRR residues could allow low-affinity interaction with a novel Avr protein variant and hence establish a weak resistance response that would be selectively advantageous to the host plants. Subsequent amino acid changes would then be selected to allow enhanced recognition and a stronger resistance phenotype. Likewise, single amino acid changes to the Avr protein could convert a fully avirulent phenotype to partial virulence, with a selective advantage to the rust, and allow subsequent selection for further changes that eliminate recognition. This model is likely to be applicable to other examples of gene-for-gene resistance based on direct R-Avr protein recognition.
The determination of the three-dimensional structures of AvrL567 proteins has also provided some clues about the possible pathogenicity-associated functions of the flax rust avirulence proteins that can now be tested experimentally. For example, a detailed study of the nucleic acid binding activity, including identification of specific binding targets, will be required to establish whether this reflects a physiologically important function during infection.
In conclusion, the results represent a significant step toward defining the resistance response at a molecular level and moving toward engineering new plant disease resistance genes to control diseases for which naturally occurring resistance is not adequate.
METHODS
Expression and Purification
Predicted mature forms of AvrL567-A, AvrL567-C, and AvrL567-D proteins (residues 24 to 150; the signal peptide was not included) were expressed in Escherichia coli as fusion proteins with N-terminal hexahistidine and ubiquitin tags (Catanzariti et al., 2004; Dodds et al., 2006). The expression plasmids were transformed into the E. coli strain BL21 (DE3) by heat shock and grown aerobically at 37°C in Luria-Bertani broth to an OD600 of 0.8 ∼ 1.0. Isopropylthio-β-galactoside was added to induce protein expression at 15°C for a further 18 to 20 h (final OD600 of 2.5 ∼ 3.0). The cells were harvested by centrifugation at 6700g for 10 min at 4°C and resuspended in one-tenth of the culture volume in buffer A (20 mM HEPES, pH 7, 300 mM NaCl, 10 mM imidazole, 1 mM phenylmethylsulfonyl fluoride, 1 mg/mL of aprotinin, 1 mg/mL of leupeptin, and 1 mg/mL of pepstatin). Cell suspensions were lysed by three freeze-thaw cycles using liquid nitrogen and in the presence of lysozyme (0.5 mg/mL) and DNase (50 units per 50 mL of lysate; Roche). Cell debris was removed by centrifugation at 15,000g for 30 min. The soluble fractions were collected and incubated with Talon resin (2 mL of prewashed resin per liter of culture; BD Biosciences) for immobilized metal affinity chromatography. After one hour of incubation on the rotating wheel at 4°C, the resin was washed with buffer A and buffer B (buffer A with 20 mM imidazole) and finally resuspended in 13 mL of buffer A containing deubiquitinating (DUB) enzyme (1:50 enzyme-to-substrate ratio) and 5 mM β-mercaptoethanol for 18 ∼ 20 h at 4°C. The cleaved AvrL567 protein was then eluted in the supernatant and further purified by size-exclusion chromatography using a Hi-Load Superdex 200 26/60 gel filtration column (GE Healthcare). Purified proteins were concentrated to ∼30 mg/mL using Amicon Ultra centrifugal filter devices with low binding Ultracel membrane (Millipore), frozen as aliquots in liquid nitrogen, and stored at −80°C. The protein concentrations were determined by measuring absorption at 280 nm, based on calculated extinction coefficient (AvrL567-A, 21,030 M−1 cm−1; AvrL567-C, 22,430 M−1 cm−1; AvrL567-D, 21,030 M−1 cm−1). The final yield was ∼10 mg of protein per liter of culture for all three proteins. The proteins were > 95% pure as determined by SDS-PAGE.
DUB was expressed as described for AvrL567 proteins, except that the overnight culture was expressed at 37°C (Catanzariti et al., 2004). The soluble fraction was incubated with Talon resin for one hour, washed with both buffer A and buffer B containing 5 mM β-mercaptoethanol, and finally eluted with buffer C (buffer A containing 150 mM imidazole and 5 mM β-mercaptoethanol). The purified DUB was dialyzed against buffer A and the protein concentrated to 5 mg/mL and stored as AvrL567 proteins.
Protein Characterization
To establish that the correct proteins were expressed, the AvrL567 proteins were characterized by mass spectrometry and N-terminal sequencing. For mass spectrometry, samples were desalted using chloroform precipitation and resuspended in 50% (v/v) acetonitrile/0.1% (v/v) acetic acid. The samples were then applied to a sinapinic acid matrix, where 1 μL aliquot of protein sample was mixed with 1∼ 4 μL of matrix. Matrix-assisted laser-desorption ionization time of flight (MALDI-TOF) was then performed using a Voyager-DE Biospectrometry workstation.
For N-terminal sequencing, proteins were transferred onto a polyvinylidene difluoride membrane (GE Healthcare) and stained with Ponceau S. The band at the correct molecular weight was excised and analyzed with a PE Applied Biosystems Procise 492 cLC protein sequencer. Both MALDI-TOF and N-terminal sequencing confirmed the identity and accurate molecular weight of the protein.
Crystallization and Crystal Structure Determination
Crystallization conditions were screened using a number of commercial and homemade sparse-matrix screens, with the hanging drop vapor diffusion and Mosquito crystallization robot (TTP LabTech). After optimization, the best crystals of AvrL567-A with the approximate size of 0.1 × 0.2 × 0.1 mm3 were obtained in 4 to 10% polyethylene glycol 8000, 0.1 M imidazole, pH 7.5 to 8.5, and 12.5 to 17.5 mM CoCl2, using a protein concentration of 25 to 35 mg/mL. The best crystals of AvrL567-D were grown in 1.26 M ammonium sulfate and 0.1 M HEPES, pH 7.5, using a protein concentration of 10 mg/mL and reached the size of 0.1 × 0.05 × 0.05 mm3.
The AvrL567-A crystals were transferred into a cryoprotectant solution containing the crystallization mother liquor containing additional 30% (v/v) 2-methyl-2,4-pentanediol and flash-cooled in liquid nitrogen. The x-ray diffraction data sets were collected in a cryostream (100 K) with a RaxisIV++ image plate detector and with CuKα radiation from a Rigaku FR-E rotating anode generator (Rigaku/MSC). The raw data sets were auto-indexed, integrated, and scaled using the HKL2000 package (Otwinowski and Minor, 1997). The crystals have the symmetry of the orthorhombic space group P212121 with unit cell lengths of a = 39.6 Å, b = 52.2 Å, and c = 70.9 Å and one molecule per asymmetric unit. Initially, a data set was collected at a maximum resolution of 2.0 Å and used to solve the structure by single anomalous dispersion phasing (Guncar et al., 2007); a data set collected at a maximum resolution of 1.4 Å was used for refinement. Briefly, the presence of Co2+ was detected using the program Hyss (Grosse-Kunstleve and Adams, 2003), which showed one Co2+ binding site with ∼100% occupancy. The Co2+ binding site was used to calculate phases with SOLVE (Terwilliger and Berendzen, 1999). Model building was performed using RESOLVE (Terwilliger, 2002), Arp/Warp (Morris et al., 2003), and MAIN (Turk, 1992), and REFMAC5 was used for refinement (Murshudov et al., 1997). Water molecules were built by Arp/Warp (Morris et al., 2003) (Table 1). The model contains residues 36 to 150 of AvrL567-A (residues 118 to 121 have poor electron density; therefore, the occupancies have been set to 0), one cobalt ion, one imidazole molecule, and 190 water molecules.
The AvrL567-D crystals were cryoprotected as for AvrL567-A. The crystals have the symmetry of the orthorhombic space group P212121 with unit cell lengths a = 39.5 Å, b = 45.8 Å, and c = 84.2 Å and one molecule per asymmetric unit. The structure was solved by molecular replacement with the program Phaser (Storoni et al., 2004), using the structure of AvrL567-A as a search model. The model was built using Arp/Warp (Morris et al., 2003) and refined as for AvrL567-A (Table 1). The model contains residues 37 to 149 of AvrL567-D and 114 water molecules.
Molecular Modeling
Molecular models of AvrL567-B, -C, -E, -F, -G, -H, -I, -J, -K, and -L were built using the AvrL567-D structure as the template with the program Modeller (Fiser and Sali, 2003). To build a molecular model of the LRR region of L5 and L6 proteins, we used the structure of internalin A (Schubert et al., 2002) as a template, as no structural information is available for any plant NBS-LRR protein, and internalin A represents the best sequence match for these proteins to any protein with known three-dimensional structure (BLAST; Altschul et al., 1990). LRR profile-based sequence alignment (Kajava and Kobe, 2002; Finn et al., 2006) of the LRR domains of the L5 and L6 proteins (residues 615 to 1251 and 607 to 1241, respectively) with internalin A (PDB ID 1O6W; Schubert et al., 2002) was generated using the Hmmer package (Eddy, 1998) and was further corrected by hand. The alignment allowed us to build models of LRR domains of L5 and L6 proteins using the program Modeller (Fiser and Sali, 2003). AvrL567-A and AvrL567-D proteins were docked to this structure using the program Zdock (Chen and Weng, 2002).
Proteolytic Assays
AvrL567-A, -C, -D, or papain (10 μM protein concentration) in 20 mM Na2HPO4/NaH2PO4 buffer, pH 5.8, were mixed with 5 mM DTT or 5 mM β-mercaptoethanol and 100 μM substrate z-Phe-Arg-pNA (Bachem) at both room temperature and 4°C. The OD410 measurements of the samples incubated at room temperature were taken at 5 min, 30 min, 2 h, and 6 h. The 4°C sample was measured after overnight incubation.
Metal Binding Studies
Purified AvrL567-A (∼8 mg/mL, 200 μL), AvrL567-C (∼10 mg/mL, 200 μL), and AvrL567-D (∼5 mg/mL, 200 μL) were mixed with 50 mM MgCl2, CaCl2, or CoCl2 or 0.1 mM ZnCl2 or NiSO4 (divalent metals most commonly found bound to proteins in the Protein Data Bank) for 3 h on ice, followed by size-exclusion chromatography using a Superdex 75 10/300 gel filtration column (GE Healthcare). The dimerized AvrL567-A fraction collected from size-exclusion chromatography was tested in the same way again in the presence of additional 0.1 mM EDTA, which dissociated the dimer.
Nucleic Acid Binding Assays
The probes were prepared as described (Kwan et al., 2003). The sequences are shown in Supplemental Table 1 online. The binding reaction with a total volume of 30 μL contained ∼0.2 pmol of 32P-labeled probe (final concentration of ∼6 nM), ∼9 μg of the AvrL567 proteins (final concentration ∼20 μM), 10 mM HEPES/NaOH, pH 7.9, 50 mM KCl, 5 mM MgCl2, 1 mM EDTA, and 5% glycerol. The binding reactions were analyzed by electrophoresis at 4°C for 150 min at 250 V on 6% native polyacrylamide gels, after 30 min of incubation on ice. The gels were analyzed using a PhosphorImager (Molecular Dynamics). Glutathione S-transferase from Schistosoma japonicum and GSF-12 (residues 1 to 95 of ZNF265; Plambeck et al., 2003) were used as negative and positive control proteins, respectively.
Site-Directed Mutagenesis and Yeast Two-Hybrid Analysis
Site-directed mutants of AvrL567 genes were constructed using the Gene-Tailor kit (Stratagene) according to the manufacturer's instructions. GAL4 DNA binding domain (BD) and transcriptional activation domain (AD) fusions to the mutant AvrL567 proteins were prepared in the pGBT9 and pGADT7 vectors, respectively (Clontech), as described (Dodds et al., 2006). L5 and L6 GBT9 and GADT7 clones were described previously (Dodds et al., 2006). Yeast transformation, His growth, and lacZ assays were performed as described in the Yeast Protocols Handbook (Clontech). Yeast proteins were extracted by the trichloroacetic acid method, separated by SDS-PAGE, and transferred to nitrocellulose membranes (Pall) by electroblotting in a Bio-Rad Mini-2D apparatus. Membranes were blocked with SuperBlock-TBST (Pierce) probed with anti-HA (Roche Molecular Systems) monoclonal antibodies followed by a blocking step with normal goat serum (Pierce) and detection with goat anti-mouse/horseradish peroxidase (Pierce). Labeling was detected with the SuperSignal West Pico chemiluminescence kit (Pierce). The mutants are unlikely to have any problems with folding because they are based on reciprocal exchanges among AvrL567-A, -C, and -D proteins; accordingly, most mutants show binding activities to L5 or L6.
Transient Expression Assays
DNA constructs encoding AvrL567 proteins lacking a signal peptide and controlled by the cauliflower mosaic virus 35S promoter were generated in the binary vector pTNotTReg as described (Dodds et al., 2004). Agrobacterium tumefaciens (GV3101-pMP90) strains containing these constructs were prepared at an OD600 of 1.0 in liquid Murashige and Skoog medium containing 200 μM acetosyringone and infiltrated into flax leaves. The flax line Bison and its near-isogenic lines containing L5, L6, and L7 have been described (Flor, 1954).
Accession Numbers
The atomic coordinates and the structure factors of the AvrL567-A and AvrL567-D structures can be found in the RCSB Protein Data Bank (Berman et al., 2000) (www.rcsb.org/pdb/) as entries 2OPC and 2QVT, respectively.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. DNA and RNA Binding by AvrL567 Proteins.
Supplemental Table 1. Pentaprobe Sequences.
Supplementary Material
Acknowledgments
We thank Chris Wood for help with N-terminal sequencing and Daigo Takemoto and David Jones for sharing unpublished data. This work was funded in part by a grant from the Australian Research Council (to B.K., P.A.A., and J.G.E.). B.K. is an Australian Research Council Federation Fellow and a National Health and Medical Research Council Honorary Research Fellow, J.K.F. was a National Health and Medical Research Council CJ Martin Fellow, and H.J.S. is a Queensland Smart State Fellow.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Boštjan Kobe (b.kobe@uq.edu.au).
Online version contains Web-only data.
References
- Abrahamsen, M.S., et al. (2004). Complete genome sequence of the apicomplexan, Cryptosporidium parvum. Science 304 441–445. [DOI] [PubMed] [Google Scholar]
- Allen, R.L., Bittner-Eddy, P.D., Grenville-Briggs, L.J., Meitz, J.C., Rehmany, A.P., Rose, L.E., and Beynon, J.L. (2004). Host-parasite coevolutionary conflict between Arabidopsis and downy mildew. Science 306 1957–1960. [DOI] [PubMed] [Google Scholar]
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215 403–410. [DOI] [PubMed] [Google Scholar]
- Anderson, P.A., Lawrence, G.J., Morrish, B.C., Ayliffe, M.A., Finnegan, E.J., and Ellis, J.G. (1997). Inactivation of the flax rust resistance gene M associated with loss of a repeated unit within the leucine-rich repeat coding region. Plant Cell 9 641–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong, M.R., et al. (2005). An ancestral oomycete locus contains late blight avirulence gene Avr3a, encoding a protein that is recognized in the host cytoplasm. Proc. Natl. Acad. Sci. USA 102 7766–7771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell, M.J., and Staskawicz, B.J. (2003). Initiation of RPS2-specified disease resistance in Arabidopsis is coupled to the AvrRpt2-directed elimination of RIN4. Cell 112 369–377. [DOI] [PubMed] [Google Scholar]
- Belkhadir, Y., Subramaniam, R., and Dangl, J.L. (2004). Plant disease resistance protein signaling: NBS-LRR proteins and their partners. Curr. Opin. Plant Biol. 7 391–399. [DOI] [PubMed] [Google Scholar]
- Berman, H.M., Bhat, T.N., Bourne, P.E., Feng, Z., Gilliland, G., Weissig, H., and Westbrook, J. (2000). The Protein Data Bank and the challenge of structural genomics. Nat. Struct. Biol. 7(Suppl): 957–959. [DOI] [PubMed] [Google Scholar]
- Catanzariti, A.M., Dodds, P.N., Lawrence, G.J., Ayliffe, M.A., and Ellis, J.G. (2006). Haustorially expressed secreted proteins from flax rust are highly enriched for avirulence elicitors. Plant Cell 18 243–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catanzariti, A.M., Soboleva, T.A., Jans, D.A., Board, P.G., and Baker, R.T. (2004). An efficient system for high-level expression and easy purification of authentic recombinant proteins. Protein Sci. 13 1331–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, R., and Weng, Z. (2002). Docking unbound proteins using shape complementarity, desolvation, and electrostatics. Proteins 47 281–294. [DOI] [PubMed] [Google Scholar]
- Chisholm, S.T., Coaker, G., Day, B., and Staskawicz, B.J. (2006). Host-microbe interactions: Shaping the evolution of the plant immune response. Cell 124 803–814. [DOI] [PubMed] [Google Scholar]
- Ciuffetti, L.M., Tuori, R.P., and Gaventa, J.M. (1997). A single gene encodes a selective toxin causal to the development of tan spot of wheat. Plant Cell 9 135–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangl, J.L., and Jones, J.D. (2001). Plant pathogens and integrated defence responses to infection. Nature 411 826–833. [DOI] [PubMed] [Google Scholar]
- Deslandes, L., Olivier, J., Peeters, N., Feng, D.X., Khounlotham, M., Boucher, C., Somssich, I., Genin, S., and Marco, Y. (2003). Physical interaction between RRS1-R, a protein conferring resistance to bacterial wilt, and PopP2, a type III effector targeted to the plant nucleus. Proc. Natl. Acad. Sci. USA 100 8024–8029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds, P.N., Lawrence, G.J., Catanzariti, A.M., Ayliffe, M.A., and Ellis, J.G. (2004). The Melampsora lini AvrL567 avirulence genes are expressed in haustoria and their products are recognized inside plant cells. Plant Cell 16 755–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds, P.N., Lawrence, G.J., Catanzariti, A.M., Teh, T., Wang, C.I., Ayliffe, M.A., Kobe, B., and Ellis, J.G. (2006). Direct protein interaction underlies gene-for-gene specificity and coevolution of the flax resistance genes and flax rust avirulence genes. Proc. Natl. Acad. Sci. USA 103 8888–8893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds, P.N., Lawrence, G.J., and Ellis, J.G. (2001. a). Six amino acid changes confined to the leucine-rich repeat beta-strand/beta-turn motif determine the difference between the P and P2 rust resistance specificities in flax. Plant Cell 13 163–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds, P.N., Lawrence, G.J., and Ellis, J.G. (2001. b). Contrasting modes of evolution acting on the complex N locus for rust resistance in flax. Plant J. 27 439–453. [DOI] [PubMed] [Google Scholar]
- Dodds, P.N., Lawrence, G.J., Pryor, A., and Ellis, J.G. (2000). Genetic analysis and evolution of plant disease resistance genes. In Molecular Plant Pathology. Annual Plant Reviews, Vol. 4, M. Dickinson and J. Beynon, eds (Sheffield, UK: Sheffield Academic Press), pp. 88–107.
- Eddy, S.R. (1998). Profile hidden Markov models. Bioinformatics 14 755–763. [DOI] [PubMed] [Google Scholar]
- Ellis, J., Dodds, P., and Pryor, T. (2000). The generation of plant disease resistance gene specificities. Trends Plant Sci. 5 373–379. [DOI] [PubMed] [Google Scholar]
- Ellis, J., Catanzariti, A.M., and Dodds, P. (2006). The problem of how fungal and oomycete avirulence proteins enter plant cells. Trends Plant Sci. 11 61–63. [DOI] [PubMed] [Google Scholar]
- Ellis, J.G., Lawrence, G.J., Luck, J.E., and Dodds, P.N. (1999). Identification of regions in alleles of the flax rust resistance gene L that determine differences in gene-for-gene specificity. Plant Cell 11 495–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn, R.D., et al. (2006). Pfam: Clans, web tools and services. Nucleic Acids Res. 34 D247–D251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiser, A., and Sali, A. (2003). Modeller: Generation and refinement of homology-based protein structure models. Methods Enzymol. 374 461–491. [DOI] [PubMed] [Google Scholar]
- Flor, H. (1971). Current status of the gene-for-gene concept. Annu. Rev. Phytopathol. 9 275–296. [Google Scholar]
- Flor, H.H. (1954). Seed-flax improvemnt. III. Flax rust. Adv. Agron. 6 152–161. [Google Scholar]
- Grosse-Kunstleve, R.W., and Adams, P.D. (2003). Substructure search procedures for macromolecular structures. Acta Crystallogr. D Biol. Crystallogr. 59 1966–1973. [DOI] [PubMed] [Google Scholar]
- Guncar, G., Wang, C.I., Forwood, J.K., Teh, T., Catanzariti, A.M., Ellis, J.G., Dodds, P.N., and Kobe, B. (2007). The use of Co2+ for crystallization and structure determination, using a conventional monochromatic X-ray source, of flax rust avirulence protein. Acta Crystallograph. Sect. F Struct. Biol. Cryst. Commun. Biol. Cryst. Commun. 63 209–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm, L., and Sander, C. (1998). Touring protein fold space with Dali/FSSP. Nucleic Acids Res. 26 316–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howles, P., Lawrence, G., Finnegan, J., McFadden, H., Ayliffe, M., Dodds, P., and Ellis, J. (2005). Autoactive alleles of the flax L6 rust resistance gene induce non-race-specific rust resistance associated with the hypersensitive response. Mol. Plant Microbe Interact. 18 570–582. [DOI] [PubMed] [Google Scholar]
- Hwang, C.F., and Williamson, V.M. (2003). Leucine-rich repeat-mediated intramolecular interactions in nematode recognition and cell death signaling by the tomato resistance protein Mi. Plant J. 34 585–593. [DOI] [PubMed] [Google Scholar]
- Hwang, S., Gou, Z., and Kuznetsov, I.B. (2007). DP-Bind: A web server for sequence-based prediction of DNA-binding residues in DNA-binding proteins. Bioinformatics 23 634–636. [DOI] [PubMed] [Google Scholar]
- Islam, M.R., and Mayo, G.M. (1990). A compendium on host genes in flax conferring resistance to flax rust. Plant Breed. 104 89–100. [Google Scholar]
- Janjusevic, R., Abramovitch, R.B., Martin, G.B., and Stebbins, C.E. (2006). A bacterial inhibitor of host programmed cell death defenses is an E3 ubiquitin ligase. Science 311 222–226. [DOI] [PubMed] [Google Scholar]
- Jia, Y., McAdams, S.A., Bryan, G.T., Hershey, H.P., and Valent, B. (2000). Direct interaction of resistance gene and avirulence gene products confers rice blast resistance. EMBO J. 19 4004–4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, D.A., and Takemoto, D. (2004). Plant innate immunity - Direct and indirect recognition of general and specific pathogen-associated molecules. Curr. Opin. Immunol. 16 48–62. [DOI] [PubMed] [Google Scholar]
- Kabsch, W., and Sander, C. (1983). Dictionary of protein secondary structure: Pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22 2577–2637. [DOI] [PubMed] [Google Scholar]
- Kajava, A.V., and Kobe, B. (2002). Assessment of the ability to model proteins with leucine-rich repeats in light of the latest structural information. Protein Sci. 11 1082–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobe, B., and Deisenhofer, J. (1995). A structural basis of the interactions between leucine-rich repeats and protein ligands. Nature 374 183–186. [DOI] [PubMed] [Google Scholar]
- Kwan, A.H., Czolij, R., Mackay, J.P., and Crossley, M. (2003). Pentaprobe: A comprehensive sequence for the one-step detection of DNA-binding activities. Nucleic Acids Res. 31 e124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahaye, T., and Bonas, U. (2001). Molecular secrets of bacterial type III effector proteins. Trends Plant Sci. 6 479–485. [DOI] [PubMed] [Google Scholar]
- Landau, M., Mayrose, I., Rosenberg, Y., Glaser, F., Martz, E., Pupko, T., and Ben-Tal, N. (2005). ConSurf 2005: The projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. 33 W299–W302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski, R.A., MacArthur, M.W., Moss, D.S., and Thornton, J.M. (1993). PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26 283–291. [Google Scholar]
- Laskowski, R.A., Watson, J.D., and Thornton, J.M. (2005). ProFunc: A server for predicting protein function from 3D structure. Nucleic Acids Res. 33 W89–W93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence, G.L., Finnegan, E.J., Ayliffe, M.A., and Ellis, J.G. (1995). The L6 gene for flax rust resistance is related to the Arabidopsis bacterial resistance gene RPS2 and the tobacco viral resistance gene N. Plant Cell 7 1195–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, C.C., Wood, M.D., Ng, K., Andersen, C.B., Liu, Y., Luginbuhl, P., Spraggon, G., and Katagiri, F. (2004). Crystal structure of the type III effector AvrB from Pseudomonas syringae. Structure 12 487–494. [DOI] [PubMed] [Google Scholar]
- Ling, H., Boudsocq, F., Woodgate, R., and Yang, W. (2001). Crystal structure of a Y-family DNA polymerase in action: A mechanism for error-prone and lesion-bypass replication. Cell 107 91–102. [DOI] [PubMed] [Google Scholar]
- Luck, J.E., Lawrence, G.J., Dodds, P.N., Shepherd, K.W., and Ellis, J.G. (2000). Regions outside of the leucine-rich repeats of flax rust resistance proteins play a role in specificity determination. Plant Cell 12 1367–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luderer, R., and Joosten, M.H. (2001). Avirulence proteins of plant pathogens: Determinants of victory and defeat. Mol. Plant Pathol. 6 355–364. [DOI] [PubMed] [Google Scholar]
- Mackey, D., Belkhadir, Y., Alonso, J.M., Ecker, J.R., and Dangl, J.L. (2003). Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2-mediated resistance. Cell 112 379–389. [DOI] [PubMed] [Google Scholar]
- Manning, V.A., Andrie, R.M., Trippe, A.F., and Ciuffetti, L.M. (2004). Ptr ToxA requires multiple motifs for complete activity. Mol. Plant Microbe Interact. 17 491–501. [DOI] [PubMed] [Google Scholar]
- Manning, V.A., and Ciuffetti, L.M. (2005). Localization of Ptr ToxA produced by Pyrenophora tritici-repentis reveals protein import into wheat mesophyll cells. Plant Cell 17 3203–3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHale, L., Tan, X., Koehl, P., and Michelmore, R.W. (2006). Plant NBS-LRR proteins: Adaptable guards. Genome Biol. 7 212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalopoulos, I., Torrance, G.M., Gilbert, D.R., and Westhead, D.R. (2004). TOPS: An enhanced database of protein structural topology. Nucleic Acids Res. 32 D251–D254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffett, P., Farnham, G., Peart, J., and Baulcombe, D.C. (2002). Interaction between domains of a plant NBS-LRR protein in disease resistance-related cell death. EMBO J. 21 4511–4519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris, R.J., Perrakis, A., and Lamzin, V.S. (2003). ARP/wARP and automatic interpretation of protein electron density maps. Methods Enzymol. 374 229–244. [DOI] [PubMed] [Google Scholar]
- Murshudov, G.N., Vagin, A.A., and Dodson, E.J. (1997). Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53 240–255. [DOI] [PubMed] [Google Scholar]
- Neuvirth, H., Raz, R., and Schreiber, G. (2004). ProMate: A structure based prediction program to identify the location of protein-protein binding sites. J. Mol. Biol. 338 181–199. [DOI] [PubMed] [Google Scholar]
- Nicholls, A., and Honig, B. (1991). A rapid finite difference algorithm, using successive over-relaxation to solve the Poisson-Boltzmann equation. J. Comput. Chem. 12 435–445. [Google Scholar]
- Nurnberger, T., Brunner, F., Kemmerling, B., and Piater, L. (2004). Innate immunity in plants and animals: Striking similarities and obvious differences. Immunol. Rev. 198 249–266. [DOI] [PubMed] [Google Scholar]
- Otwinowski, Z., and Minor, W. (1997). Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276 307–326. [DOI] [PubMed] [Google Scholar]
- Pal, D., and Eisenberg, D. (2005). Inference of protein function from protein structure. Structure 13 121–130. [DOI] [PubMed] [Google Scholar]
- Plambeck, C.A., Kwan, A.H., Adams, D.J., Westman, B.J., van der Weyden, L., Medcalf, R.L., Morris, B.J., and Mackay, J.P. (2003). The structure of the zinc finger domain from human splicing factor ZNF265 fold. J. Biol. Chem. 278 22805–22811. [DOI] [PubMed] [Google Scholar]
- Rairdan, G.J., and Moffett, P. (2006). Distinct domains in the ARC region of the potato resistance protein Rx mediate LRR binding and inhibition of activation. Plant Cell 18 2082–2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehmany, A.P., Gordon, A., Rose, L.E., Allen, R.L., Armstrong, M.R., Whisson, S.C., Kamoun, S., Tyler, B.M., Birch, P.R., and Beynon, J.L. (2005). Differential recognition of highly divergent downy mildew avirulence gene alleles by RPP1 resistance genes from two Arabidopsis lines. Plant Cell 17 1839–1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooney, H.C., Van't Klooster, J.W., van der Hoorn, R.A., Joosten, M.H., Jones, J.D., and de Wit, P.J. (2005). Cladosporium Avr2 inhibits tomato Rcr3 protease required for Cf-2-dependent disease resistance. Science. 308 1783–1786. [DOI] [PubMed] [Google Scholar]
- Sarma, G.N., Manning, V.A., Ciuffetti, L.M., and Karplus, P.A. (2005). Structure of Ptr ToxA: An RGD-containing host-selective toxin from Pyrenophora tritici-repentis. Plant Cell 17 3190–3202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert, W.D., Urbanke, C., Ziehm, T., Beier, V., Machner, M.P., Domann, E., Wehland, J., Chakraborty, T., and Heinz, D.W. (2002). Structure of internalin, a major invasion protein of Listeria monocytogenes, in complex with its human receptor E-cadherin. Cell 111 825–836. [DOI] [PubMed] [Google Scholar]
- Shao, F., Golstein, C., Ade, J., Stoutemyer, M., Dixon, J.E., and Innes, R.W. (2003). Cleavage of Arabidopsis PBS1 by a bacterial type III effector. Science 301 1230–1233. [DOI] [PubMed] [Google Scholar]
- Shcherbakova, P.V., Bebenek, K., and Kunkel, T.A. (2003). Functions of eukaryotic DNA polymerases. Sci. Aging Knowledge Environ. 2003: RE3. [DOI] [PubMed]
- Shen, Q.H., Zhou, F., Bieri, S., Haizel, T., Shirasu, K., and Schulze-Lefert, P. (2003). Recognition specificity and RAR1/SGT1 dependence in barley Mla disease resistance genes to the powdery mildew fungus. Plant Cell 15 732–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shindyalov, I.N., and Bourne, P.E. (1998). Protein structure alignment by incremental combinatorial extension (CE) of the optimal path. Protein Eng. 11 739–747. [DOI] [PubMed] [Google Scholar]
- Storoni, L.C., McCoy, A.J., and Read, R.J. (2004). Likelihood-enhanced fast rotation functions. Acta Crystallogr. D Biol. Crystallogr. 60 432–438. [DOI] [PubMed] [Google Scholar]
- Takken, F.L., Albrecht, M., and Tameling, W.I. (2006). Resistance proteins: Molecular switches of plant defence. Curr. Opin. Plant Biol. 9 383–390. [DOI] [PubMed] [Google Scholar]
- Tameling, W.I., Vossen, J.H., Albrecht, M., Lengauer, T., Berden, J.A., Haring, M.A., Cornelissen, B.J., and Takken, F.L. (2006). Mutations in the NB-ARC domain of I-2 that impair ATP hydrolysis cause autoactivation. Plant Physiol. 140 1233–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger, T.C. (2002). Automated structure solution, density modification and model building. Acta Crystallogr. D Biol. Crystallogr. 58 1937–1940. [DOI] [PubMed] [Google Scholar]
- Terwilliger, T.C., and Berendzen, J. (1999). Automated MAD and MIR structure solution. Acta Crystallogr. D Biol. Crystallogr. 55 849–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya, Y., Kinoshita, K., and Nakamura, H. (2005). PreDs: A server for predicting dsDNA-binding site on protein molecular surfaces. Bioinformatics 21 1721–1723. [DOI] [PubMed] [Google Scholar]
- Turk, D. (1992). Weiterentwicklung eines Programms fur Molekulgraphik und Elektrondichte-Manipulation und seine Anwendung auf verschiedene Protein-Strukturaufklarungen. PhD dissertation (Muenchen, Germany: Technische Universitat).
- Ueda, H., Yamaguchi, Y., and Sano, H. (2006). Direct interaction between the tobacco mosaic virus helicase domain and the ATP-bound resistance protein, N factor during the hypersensitive response in tobacco plants. Plant Mol. Biol. 61 31–45. [DOI] [PubMed] [Google Scholar]
- Vaguine, A.A., Richelle, J., and Wodak, S.J. (1999). SFCHECK: A unified set of procedures for evaluating the quality of macromolecular structure-factor data and their agreement with the atomic model. Acta Crystallogr. D Biol. Crystallogr. 55 191–205. [DOI] [PubMed] [Google Scholar]
- van den Burg, H.A., Westerink, N., Francoijs, K.J., Roth, R., Woestenenk, E., Boeren, S., de Wit, P.J., Joosten, M.H., and Vervoort, J. (2003). Natural disulfide bond-disrupted mutants of AVR4 of the tomato pathogen Cladosporium fulvum are sensitive to proteolysis, circumvent Cf-4-mediated resistance, but retain their chitin binding ability. J. Biol. Chem. 278 27340–27346. [DOI] [PubMed] [Google Scholar]
- van den Hooven, H.W., van den Burg, H.A., Vossen, P., Boeren, S., de Wit, P.J., and Vervoort, J. (2001). Disulfide bond structure of the AVR9 elicitor of the fungal tomato pathogen Cladosporium fulvum: Evidence for a cystine knot. Biochemistry 40 3458–3466. [DOI] [PubMed] [Google Scholar]
- van't Slot, K.A., van den Burg, H.A., Kloks, C.P., Hilbers, C.W., Knogge, W., and Papavoine, C.H. (2003). Solution structure of the plant disease resistance-triggering protein NIP1 from the fungus Rhynchosporium secalis shows a novel beta-sheet fold. J. Biol. Chem. 278 45730–45736. [DOI] [PubMed] [Google Scholar]
- Wulf, J., Pascuzzi, P.E., Fahmy, A., Martin, G.B., and Nicholson, L.K. (2004). The solution structure of type III effector protein AvrPto reveals conformational and dynamic features important for plant pathogenesis. Structure 12 1257–1268. [DOI] [PubMed] [Google Scholar]
- Zhu, M., Shao, F., Innes, R.W., Dixon, J.E., and Xu, Z. (2004). The crystal structure of Pseudomonas avirulence protein AvrPphB: A papain-like fold with a distinct substrate-binding site. Proc. Natl. Acad. Sci. USA 101 302–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.