Abstract
Protein 6b, encoded by T-DNA from the pathogen Agrobacterium tumefaciens, stimulates the plant hormone–independent division of cells in culture in vitro and induces aberrant cell growth and the ectopic expression of various genes, including genes related to cell division and meristem-related class 1 KNOX homeobox genes, in 6b-expressing transgenic Arabidopsis thaliana and Nicotiana tabacum plants. Protein 6b is found in nuclei and binds to several plant nuclear proteins. Here, we report that 6b binds specifically to histone H3 in vitro but not to other core histones. Analysis by bimolecular fluorescence complementation revealed an interaction in vivo between 6b and histone H3. We recovered 6b from a chromatin fraction from 6b-expressing plant cells. A supercoiling assay and digestion with micrococcal nuclease indicated that 6b acts as a histone chaperone with the ability to mediate formation of nucleosomes in vitro. Mutant 6b, lacking the C-terminal region that is required for cell division–stimulating activity and interaction with histone H3, was deficient in histone chaperone activity. Our results suggest a relationship between alterations in nucleosome structure and the expression of growth-regulating genes on the one hand and the induction of aberrant cell proliferation on the other.
INTRODUCTION
Cells of Agrobacterium tumefaciens that harbor a Ti plasmid induce the formation of tumors, known as crown galls, on many dicotyledonous plants. Upon infection by the bacterium, a specific region of the Ti plasmid (T-DNA) is transferred to the plant cell and is integrated into the nuclear chromosomal DNA. Cells with chromosomally integrated T-DNA can divide autonomously to generate tumors, which appear to be a consequence, for the most part, of the expression of genes that are responsible for the biosynthesis of auxin (aux1/iaaM/tms1 and aux2/iaaH/tms2) and cytokinin (cyt/ipt/tmr; Morris, 1986). In addition to these genes, a gene designated 6b, which is localized at the tml locus (Garfinkel et al., 1981; Willmitzer et al., 1983) and has been found in the T-DNA of all strains of A. tumefaciens and Agrobacterium vitis examined to date (Otten and De Ruffray, 1994), appears to play a role in the proliferation of plant cells. This gene modifies the morphology of crown galls but is not required for their formation (Garfinkel et al., 1981; Tinland et al., 1992), and it induces callus formation on tobacco (Nicotiana tabacum) leaf discs in the absence of exogenous auxin and cytokinin (Wabiko and Minemura, 1996; Kitakura et al., 2002).
Various hypotheses have been proposed to explain the effects of 6b on the growth of plant cells (Leemans et al., 1982; Ream et al., 1983; Hooykaas et al., 1988; Tinland et al., 1992; Wabiko and Minemura, 1996). This gene modulates the inductive effects of cytokinins on shoot development (Spanier et al., 1989); it interferes with the induction and elongation of roots in an auxin-dependent rolABC-based rooting assay, suggesting that 6b might reduce the effect of high levels of auxin to maintain cells in an undifferentiated state (Tinland et al., 1990); and it enhances the effects of both auxin and cytokinin on crown gall formation (Canaday et al., 1992). There are some discrepancies among previously reported results, but it is generally accepted that 6b affects the proliferation of cells and the development of shoots by modulating the actions of cytokinin and/or auxin. In addition, it was reported recently that 6b modulates the metabolism of phenolic compounds (Gális et al., 2002; Kakiuchi et al., 2006) and induces cell expansion by increasing osmolyte concentrations (Clément et al., 2006).
The predicted amino acid sequence of 6b is somewhat similar to those of a number of proteins encoded by other genes, such as rolB and ORF13 in Ri plasmids, which also cause aberrant growth and the abnormal morphology of both roots and shoots (Spena et al., 1987; Levesque et al., 1988; Stieger et al., 2004). The product of ORF13 has a retinoblastoma binding motif (LXCXE) and interacts with maize (Zea mays) Rb in vitro (Stieger et al., 2004). RolB, which includes a Tyr phosphatase motif (CX5R), has phosphatase activity (Filippini et al., 1996). Neither motif is found in 6b, and the way in which 6b acts at the molecular level remains to be determined. Recently, we identified three tobacco nuclear proteins that can interact with 6b. The interaction of 6b with one of these proteins, the transcription factor–like protein Nt SIP1, facilitates the nuclear localization of 6b in plant cells (Kitakura et al., 2002), which is essential for the 6b-induced abnormal growth of cells (Terakura et al., 2006). We showed that expression of the 6b gene causes the ectopic expression of variety of genes, such as class I KNOX, CUC, and cell cycle–related genes, in tobacco and Arabidopsis thaliana (Terakura et al., 2006). These observations suggest possible roles for 6b in the nucleus. Here, we show that 6b interacts with the histone fold of histone H3 and that it has histone chaperone–like activity, which is essential for the hormone-independent growth of plant cells that is mediated by 6b. Our results suggest the importance of putative 6b-induced alterations in the structure of plant chromatin in the unregulated growth of cells.
RESULTS
Protein 6b Interacts with the Histone Fold of Histone H3 in Vitro
The 6b protein includes a cluster of acidic amino acid residues near its C terminus (see Supplemental Figure 1A online). This acidic region is required both for the interaction of 6b with the tobacco nuclear protein Nt SIP1 and for the induction by 6b of the formation of calli on hormone-free medium (Figure 1A; Kitakura et al., 2002). To identify tobacco proteins that might bind to regions of 6b other than the acidic region, we screened a tobacco cDNA library (1.5 × 106 independent clones) in a yeast two-hybrid system, using the 6bΔA sequence (see Supplemental Figure 1A online) that lacks the acidic region. We identified four positive clones (see Supplemental Table 1 online). Two of these cDNA clones (clones 1 and 2) encoded partial sequences of members of the histone H3 family. We isolated tobacco cDNA clones that encoded entire coding regions corresponding to clones 1 and 2. The amino acid sequences that we deduced from these clones revealed that amino acid sequences corresponding to clones 1 and 2 were identical to those of histone H3.1 and H3.2 of Arabidopsis, respectively (see Supplemental Table 1 and Supplemental Figure 2 online; Waterborg and Robertson, 1996; Johnson et al., 2004). Clone 3 encoded an amino acid sequence that was somewhat similar to that of the histone fold domain of histone H2B, and clone 4 encoded a protein with no significant homology to previously characterized sequences. In this study, we performed further experiments with the cDNA of clone 2 since it has been suggested that H3.2 is equivalent to replication-independent (replacement) histone H3 that has been found in animal cells (Johnson et al., 2004).
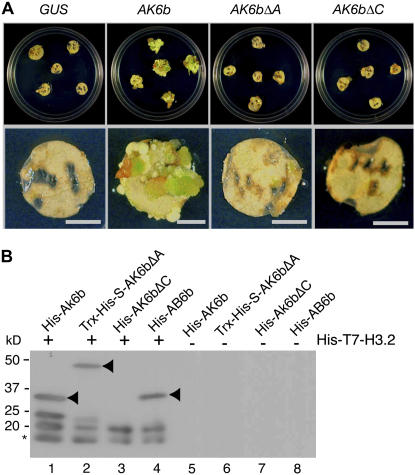
Figure 1.
Mitogenic Activity of 6b and Its Interaction with Histone H3.
(A) Callus-formation assay. DNA constructs for recombinant proteins of wild-type AK6b and mutant AK6bΔA and AK6bΔC, as described in Methods and in Supplemental Figure 1 online, were linked to the 35S promoter of Cauliflower mosaic virus, and each 6b construct and a control β-glucuronidase (GUS) gene construct were introduced into cells of tobacco leaf discs by a Ti plasmid vector system. The leaf discs were incubated on agar plates that contained selective medium without cytokinin and auxin (see Methods). Bars = 5 mm.
(B) Binding of 6b and its derivatives to histone H3.2. Recombinant His-T7-histone H3.2 protein was incubated with His-AK6b (lanes 1 and 5), Trx-His-S-AK6bΔA (lanes 2 and 6), His-AK6bΔC (lanes 3 and 7), and His-AB6b (lanes 4 and 8), and protein complexes were immunoprecipitated with T7-specific antibodies as described in the text. The recovered complexes were subjected to SDS-PAGE (10.5% acrylamide) and protein gel blot analysis with His-specific antibodies. Proteins of 6b and its derivatives are indicated by arrowheads. Some 6b protein was detected as two forms (lanes 1 and 2), one of which might have been a degradation product. His-T7-histone H3.2 was also detected as two forms. The rapidly migrating proteins, indicated by an asterisk, might also represent degradation products.
We performed coimmunoprecipitation experiments with His epitope-tagged 6b (His-6b) and His epitope-plus T7 epitope-tagged tobacco histone H3.2 (His-T7-histone H3.2) to examine their interactions in vitro. In these binding assays, we prepared two kinds of recombinant 6b protein: one from A. tumefaciens AKE10 (designated His-AK6b) and one from A. vitis AB4 (designated His-AB6b; see Supplemental Figure 1B online) because these 6b proteins, from two different bacterial sources, are both able to induce abnormal morphology in tobacco and Arabidopsis plants (Wabiko and Minemura, 1996; Helfer et al., 2003; Terakura et al., 2006). We also generated Trx-His-S epitope-tagged AK6bΔA (Trx-His-S-AK6bΔA) and His-AK6bΔC proteins that lacked the acidic region and the C-terminal region of 6b, respectively (see Supplemental Figure 1A online). The purity of the recombinant tagged proteins is shown in Supplemental Figure 1B online. Even when His-AK6bΔC was expressed under the control of the 35S promoter in cells of tobacco leaf discs, it was unable to support plant hormone–independent growth (Figure 1A).
Using these above-mentioned recombinant proteins, we examined the possible association of 6b with histone H3 in vitro. The purified His-T7-histone H3.2 protein was incubated with His-AK6b, Trx-His-S-AK6bΔA, His-AK6bΔC, and His-AB6b, and then protein complexes were precipitated with T7-specific antibodies. Analysis of the various immunocomplexes showed that AK6b, AK6bΔA, and AB6b bound histone H3.2 (Figure 1B, arrowheads in lanes 1, 2, and 4), while AK6bΔC did not (lane 3). When wild-type and mutant 6b proteins alone were incubated with T7-specific antibodies, no positive signal was detected (Figure 1B, lanes 5 to 8). These results indicated that the C-terminal region of AK6b was required both for the binding to histone H3 and for the hormone-independent growth of tobacco cells that was mediated by 6b. Arabidopsis histone H3.1 also bound to AK6b and AB6b proteins in vitro (see Supplemental Figure 3 online).
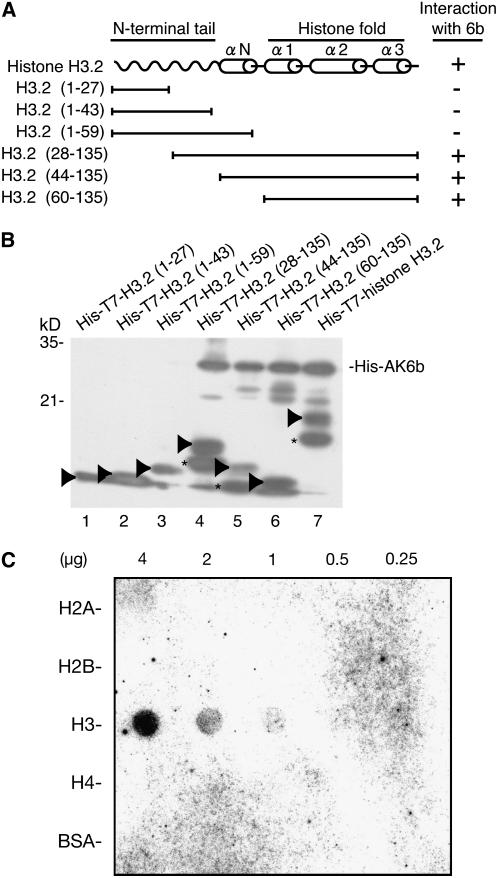
Next, we attempted to identify the region(s) in histone H3.2 that might be required for the interaction with 6b in vitro. Core histones (H2A, H2B, H3, and H4) consist of an N-terminal tail and a C-terminal histone fold that includes three α helices (α1, α2, and α3). The N-terminal tail is a prime target for the posttranslational modification of histones, and the histone fold domain in the C terminus is involved in interactions with other core histones during the formation of nucleosomes (Luger et al., 1997). We generated various truncated derivatives of histone H3.2, which were tagged with the His-T7 epitope, in Escherichia coli cells (Figure 2A). Analysis of the immunocomplexes formed by His-AK6b and these truncated histone H3.2 proteins showed that AK6b protein bound to histone H3.2(28-135), histone H3.2(44-135), and histone H3.2(60-135) (Figure 2B), all of which contained the histone fold domain but not the region that contained the histone tail and the N-terminal α helix. These results demonstrated that 6b interacted with the histone fold of histone H3.2. We also examined the specificity of the interaction between 6b and core histones in vitro. Protein blot analysis, with recombinant mammalian histones H2A, H2B, H3, and H4 and radiolabeled 6b, showed that 6b interacted with histone H3 specifically (Figure 2C).
Figure 2.
Protein 6b Interacts Specifically with the Histone Fold of Histone H3.
(A) Schematic representation of histone H3.2 and the truncated derivatives used in the binding assay.
(B) Binding of 6b to the histone fold. His-T7 epitope-tagged deleted variants of histone H3.2 and His-AK6b, as indicated in (A), were incubated together, and complexes were immunoprecipitated with T7-specific antibodies. Recovered complexes were subjected to SDS-PAGE (10.5% acrylamide) and immunoblotted with His-specific antibodies. Arrowheads indicate positions of histone H3.2 (lane 7) and its various truncated derivatives (lanes 1 to 6). The proteins indicated by asterisks might represent degradation products of the recombinant histone H3.2 proteins.
(C) Protein gel blot assay of the interaction of core histones with radiolabeled AK6b. The amount of protein in each spot is indicated above the panel. Blotting analysis was performed as described in the text.
Protein 6b Interacts with Histone H3 in Chromatin in Vivo
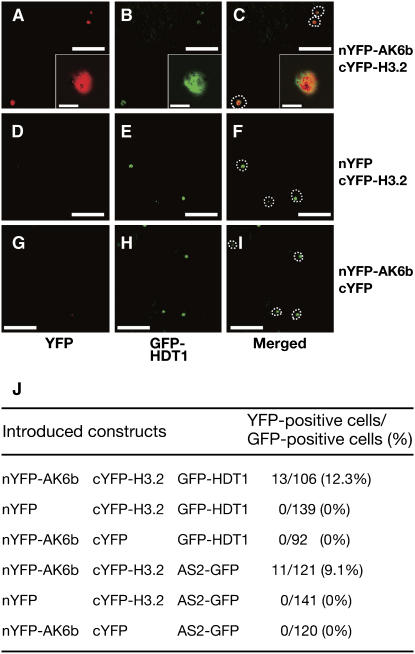
To examine interactions in vivo, we investigated the possible interaction between AK6b and histone H3.2 in plant cells by bimolecular fluorescence complementation (BiFC; Hu et al., 2002). We made two DNA constructs: in one, the DNA that encoded the N-terminal half of yellow fluorescent protein (YFP) was fused to the DNA that encoded AK6b (nYFP-AK6b); in the other, the DNA encoding the C-terminal half of YFP was fused to the DNA that encoded histone H3.2 (cYFP-H3.2). We inserted each construct separately into the binary vector plasmid and transformed Agrobacterium separately with each vector construct. We introduced both Agrobacterium strains into leaf cells of Nicotiana benthamiana and examined the fluorescence due to YFP. As shown in Figure 3, we detected fluorescence due to YFP in leaf epidermal cells of N. benthamiana that coexpressed nYFP-AK6b and cYFP-H3.2, while no fluorescence was detected in cells that coexpressed either nYFP plus cYFP-H3.2 or nYFP-AK6b plus cYFP. These results demonstrated that AK6b interacted with histone H3 in vivo.
Figure 3.
BiFC Analysis of the Interaction between 6b and Histone H3 in Leaf Epidermis of N. benthamiana.
nYFP-AK6b was coexpressed with cYFP-H3 ([A] to [C]), nYFP was coexpressed with cYFP-H3 ([D] to [F]), and nYFP-AK6b was coexpressed with cYFP ([G] to [I]). Green fluorescent protein (GFP)-HDT1 and AS2-GFP were included as internal controls. YFP fluorescence due to BiFC is shown ([A], [D], and [G]); GFP fluorescence due to GFP-HDT1 and AS2-GFP is shown ([B], [E], [H], and [J]); and merged views of YFP and GFP fluorescence are shown ([C], [F], and [I]). Insets in (A) to (C) show magnified views of (A) to (C), respectively. The profiles of nuclei are indicated by a dashed line in (C), (F), and (I). Bars = 3 μm in insets in (A) to (C) and 20 μm in all other panels.
(J) The ratio of YFP-positive cells to GFP-positive cells.
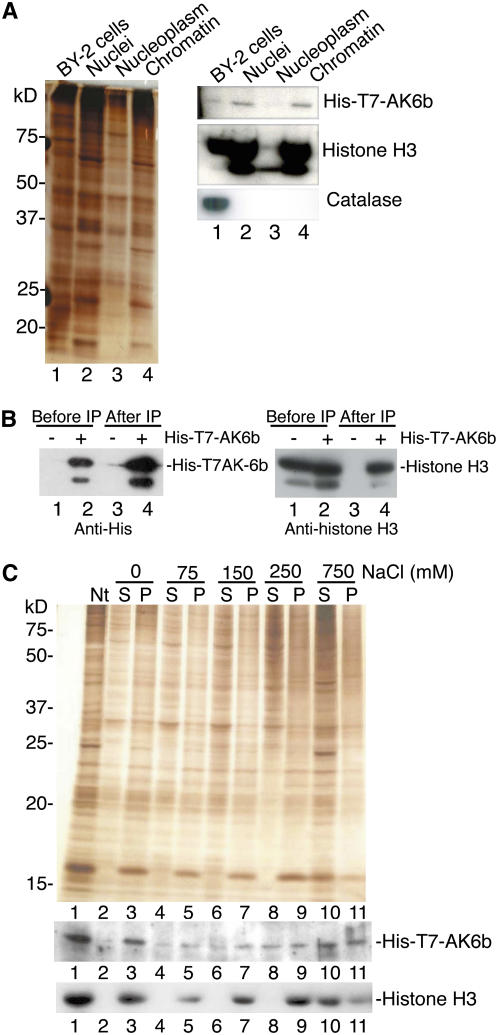
Since most histone H3 molecules in nuclei are found in association with the chromatin, the results described in the preceding paragraph suggest that 6b is associated with histone H3 in chromatin. To examine this possibility, we isolated nuclei from tobacco cultured BY-2 cells that produced His-T7-AK6b and disrupted the nuclei in a hypotonic buffer. Then we separated the chromatin fraction from the nucleoplasmic fraction by centrifugation. We performed protein gel blot analysis of the nuclear, chromatin, and nucleoplasmic fractions with histone H3–specific and T7-specific antibodies (Figure 4A). In addition, we used catalase-specific antibodies to detect catalase as a marker in the cytoplasmic fraction. Our results revealed that the nuclear and chromatin fractions contained both histone H3 and His-T7-AK6b, while the nucleoplasmic fraction did not (Figure 4A), indicating that 6b can associate with tobacco chromatin.
Figure 4.
Interaction of 6b with Histone H3 in the Chromatin of Tobacco Cells.
(A) Detection of 6b in the chromatin fraction. Protein extracts were prepared from BY-2 cells that expressed His-T7-AK6b (lane 1), from isolated nuclei (lane 2), from the nucleoplasmic fraction (lane 3), and from the chromatin fraction (lane 4). These extracts were fractionated by SDS-PAGE (10.5% acrylamide). Panels show silver staining (left) and immunoblotting analysis (right) with T7-specific (top), histone H3–specific (middle), and catalase-specific (bottom) antibodies.
(B) Association of 6b with histone H3 in nuclei. Nuclei were isolated from wild-type tobacco BY-2 cells (−; lanes 1 and 3) and from cells that expressed His-T7-AK6b (+; lanes 2 and 4), and then isolated nuclei were treated with MNase as described in the text. Immunoprecipitates (IP) were recovered from the nuclear fraction with T7-specific antibodies and immunoblotted with His-specific (left) and histone H3–specific (right) antibodies.
(C) Release of His-T7-AK6b from chromatin by NaCl. Nuclei (Nt) (lane 1) were treated with indicated concentrations of NaCl as described in the text. Pellets (P) were separated from supernatants (S) by centrifugation, and equivalent amounts of protein were subjected to SDS-PAGE (10 to 20% acrylamide) and protein gel blot analysis. Results of silver staining (top) and of immunoblotting with T7-specific (middle) and histone H3–specific (bottom) antibodies are shown.
We examined whether 6b might interact with nucleosomal histone H3. We treated isolated nuclei from transgenic tobacco BY-2 cells that expressed His-T7-AK6b with micrococcal nuclease (MNase), and then we immunoprecipitated complexes by adding T7-specific antibodies to the MNase-treated nuclear fraction. Protein gel blotting with histone H3–specific antibodies showed that the immunocomplex contained endogenous histone H3 of tobacco (Figure 4B), revealing a physical interaction between AK6b and histone H3 in tobacco cells.
We compared the affinity of His-T7-AK6b for chromatin to that of histone H3 by treating chromatin fractions with various concentrations of NaCl and examining the amounts of these proteins in the NaCl-soluble (supernatant) and NaCl-insoluble (pellet) fractions. Protein gel blot analysis showed that, as the concentration of NaCl increased, the amount of His-T7-AK6b and the amount of histone H3 released from the chromatin increased gradually (Figure 4C). However, we noted a difference in the amount of each protein released from the chromatin: endogenous histone H3 was not released to any significant extent from the chromatin at 75, 150, and 250 mM NaCl, even though some His-T7-AK6b was clearly released from the chromatin at these concentrations of NaCl. At 750 mM NaCl, a significant amount of His-T7-6b and of histone H3 remained bound to the chromatin fraction. These results suggest that individual molecules of the 6b protein might bind to H3 in the chromatin with somewhat different affinities.
Protein 6b Has Histone Chaperone–Like Activity
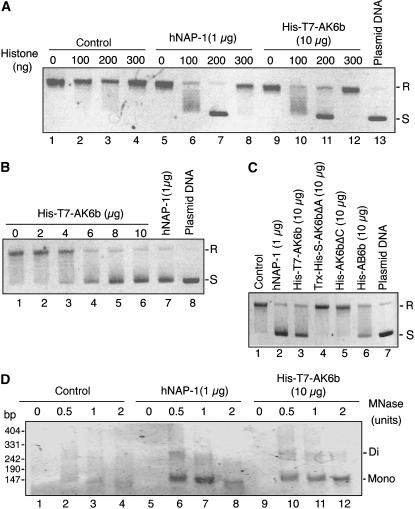
The physical association of 6b with the histone fold of histone H3 and the presence of the acidic region in 6b led us to examine 6b for histone chaperone activity since some histone chaperones, such as HIRA (histone regulation) and ASF1 (antisilencing function1), bind to the histone fold domains of core histones (Munakata et al., 2000; Ray-Gallet et al., 2002); furthermore, histone chaperones, such as NAP-1 (nucleosome assembly protein-1) (Ishimi and Kikuchi, 1991), nucleophosmin/B23 (Okuwaki et al., 2001), yeast FK506bp (Kuzuhara and Horikoshi, 2004), and nucleolin (Angelov et al., 2006), contain clusters of acidic amino acid residues. Histone chaperones enhance the formation of nucleosomes in vitro, and their activity can be detected by DNA supercoiling assays, which measure the conversion of relaxed and closed-circular DNA to negatively supercoiled and closed-circular DNA, and treatment with MNase (Laskey et al., 1978; Ishimi et al., 1983).
To examine the putative histone chaperone activity of 6b, we first performed supercoiling assays with relaxed and closed-circular DNA, recombinant 6b (or human NAP-1 [hNAP-1]) that had been purified from E. coli, and core histones prepared from HeLa cells. Figures 5A and 5C show that both AK6b and AB6b had supercoiling activity, with maximum detectable activity in the presence of 10 μg of purified His-T7-AK6b or His-AB6b and 200 ng of core histones. When we used 1 μg of human hNAP-1 protein, which has been shown to have histone chaperone activity (Fujii-Nakata et al., 1992), maximum activity was also detected with 200 ng of core histones. The molar ratio of core histones to plasmid DNA under our conditions was 1:1, which corresponds to the normal ratio for detection of maximal supercoiling activity. When we increased the amount of core histones in the assay to 300 ng, we detected only very low activity due to hNAP-1 (lane 8 in Figure 5A). We can reasonably assume that the activity that we detected might be relevant to normal reactions mediated by a histone chaperone–like protein (Fujii-Nakata et al., 1992; Kuzuhara and Horikoshi, 2004). However, we had to add ∼15 times more His-T7-AK6b than hNAP-1 for maximum conversion to negative supercoils (Figure 5B). As shown in Figure 5C, the mutant derivatives of AK6bΔA and AK6bΔC, which lacked tumorigenic activity (Figure 1B), had no histone chaperone–like activity.
Figure 5.
Histone Chaperone–Like Activity of 6b in Vitro.
(A) Supercoiling activity of 6b in the presence of core histones. Supercoiling activities of hNAP-1 and 6b for conversion of relaxed and closed-circular plasmid DNA to negatively supercoiled and closed-circular DNA were examined in the presence and in the absence of various amounts of core histones as described in Methods. Reactions were performed without a histone chaperone (Control; lanes 1 to 4), with hNAP-1 (lanes 5 to 8), and with His-T7-AK6b (lanes 9 to 12). After deproteinization, plasmid DNA was subjected to agarose gel electrophoresis (1% agarose) and visualized by staining with ethidium bromide. R and S represent positions of relaxed, closed-circular and negatively supercoiled, closed-circular plasmid DNAs, respectively.
(B) Stoichiometry of the levels of His-T7-AK6b and core histones in the supercoiling reaction. Numbers above the panel represent amounts of His-T7-6b proteins included in the reaction mixture.
(C) Supercoiling activities of the His-AB6b protein and mutant derivatives of AK6b (Trx-His-S-AK6bΔA and His-AK6bΔC). Control, no histone chaperone.
(D) Detection of nucleosomal arrays assembled in the presence of 6b. Complexes that might have been generated without (Control; lanes 1 to 4) or with hNAP-1 (lanes 5 to 8) and with His-T7-AK6b (lanes 9 to 12) were digested with MNase. Mono and Di refer to lengths (in base pairs) of polynucleotide chains covered with mono- and dinucleosomes, respectively.
To examine the possible formation of nucleosome-related structures, we digested the plasmid DNA, after the supercoiling reaction, with MNase. As shown in Figure 5D, DNA fragments of distinct lengths (∼140 and 280 bp) were generated, indicating that nucleosome-related structures had been formed in the presence of either AK6b or hNAP-1 and core histones. Our observations indicated that 6b had histone chaperone–like activity in vitro but that the activity of 6b was lower than that of hNAP-1 in our assay system.
6b Affects Levels of Transcripts of a Variety of Genes, Including IAA Genes in Arabidopsis Plants
We have reported that genes involved in the differentiation of organs (such as class 1 KNOX and CUC genes) and in the proliferation of cells (such as the gene for cyclin B) are expressed ectopically in the 6b-transgenic Arabidopsis and tobacco plants (Terakura et al., 2006). We performed microarray analyses using 6b-transgenic and nontransgenic Arabidopsis plants and found that expression of a number of auxin-inducible genes, including AUX/IAA and ACS (1-aminocyclopropane-1-carboxylate synthase gene that is involved in ethylene biosynthesis in response to auxin), was repressed in the 6b-transgenic Arabidopsis plants. We investigated how rapidly 6b can repress levels of transcripts of IAA3/SHY2 and IAA6 genes using a dexamethasone (DEX)-inducible expression system in Arabidopsis. The result showed that levels of transcripts of both IAA genes decreased as early as 6 h after induction of the transcription of the 6b gene by the addition of DEX (see Supplemental Figure 4 online). Thus, 6b appears to influence the transcription of these genes.
To investigate the relationship between the expression of the above-mentioned genes and the functions of 6b, we investigated patterns of methylation and acetylation of histones in putative promoter regions of the IAA3 and IAA6 genes in 6b-expressing and untransformed Arabidopsis plants. However, we failed to detect any differences in such modification of histones between the two types of plants (data not shown).
DISCUSSION
Protein 6b Acts as a Histone Chaperone, Influencing the Transcription of Certain Plant Genes
The results obtained in this study demonstrate that 6b protein has the ability to bind to histone H3 both in vitro and in vivo (Figures 1B and 3), to convert relaxed and closed-circular DNA to supercoiled DNA in a supercoiling assay in vitro (Figures 5A and 5B), and to form nucleosomes on DNA molecules (Figure 5D). These results indicate that 6b has the ability to mediate the assembly of core histones on DNA for the formation of nucleosomes in vitro. These characteristics of 6b are similar to those of previously identified histone chaperones in animal and yeast cells (Loyola and Almouzni, 2004), such as NAP-1, Asf1, HIRA, and TAF-I (template activating factor-I; Okuwaki and Nagata, 1998), suggesting that 6b acts as a histone H3 chaperone in plant cells. We found that the 6bΔA and 6bΔC mutant proteins (see Supplemental Figure 1A online), which were unable to support plant hormone–independent growth (Figure 1A), did not bind to histone H3 (Figure 1B) and did not have supercoiling activity (Figure 5C). These results suggest that the abnormal growth of calli induced by 6b is dependent on the histone chaperone–like activity of 6b. Although plant homologs of NAP1 (Dong et al., 2004; Galichet and Gruissem, 2006), Asf1 (Silljé and Nigg, 2001; Ehsan et al., 2004), and HIRA (Phelps-Durr et al., 2005) have been identified, activities of these homologs as histone chaperones remain to be demonstrated.
The histone chaperones of animal and yeast cells mediate, both in vitro and in vivo, the disassembly of nucleosomes and the assembly of core histones, acting in concert with chromatin-remodeling complexes and histone-modifying enzymes (Workman, 2006; Li et al., 2007). In yeast, Asf1, which is a histone H3/H4 chaperone, mediates the disassembly of nucleosomes from PHO5 and PHO8 promoters during the activation of the transcription initiation (Adkins et al., 2004). Recently, Schwabish and Struhl (2006) reported that Asf1 associates with both the promoters and the coding regions of transcriptionally active genes and mediates eviction (displacement) of histone H3 and deposition of the same histones during transcriptional elongation by RNA polymerase II. They proposed that Asf1 functions as an elongation factor to disassemble and reassemble histones during transcriptional elongation. With respect to the role of 6b in vivo, a recent report about histone chaperone TAF-I, which appears to regulate transcription either positively or negatively in a gene-specific manner, is particularly relevant (Kato et al., 2006). TAF-I bound to core histones and remodeled the chromatin in vitro. It also stimulated transcription of a number of endogenous genes and of a model reporter gene that had been integrated into the chromatin in a histone chaperone–dependent manner in vivo. Thus, these histone chaperones appear to stimulate transcription by RNA polymerase II and to mediate the turnover of nucleosomes (eviction and deposition of histones) during the elongation of RNA, suggesting a close relationship between the transcriptional elongation and the turnover of nucleosomes.
These results have shown that protein 6b appears to rapidly influence transcription of IAA3/SHY2 and IAA6 (see Supplemental Figure 4 online), although it remains to be determined whether this role of 6b is direct or indirect. The histone chaperone–like activity of 6b, detected in this study, suggests that 6b might alter the patterns of expression of these genes through its activity. Histone chaperone Asf1, which participates in the eviction and deposition of histone H3 during transcriptional elongation by RNA polymerase II (Schwabish and Struhl, 2006), is copurified with H3 and H4 (Tyler et al., 1999). Our observation that the 6b protein is associated with chromatin in 6b-transformed tobacco BY-2 cells (Figure 4) is consistent with the above hypothesis. It is important, now, to identify genes whose transcription is directly affected by 6b and to the role of 6b in the stimulation of transcription of these genes.
Although levels of transcripts of IAA3/SHY2 and IAA6 clearly decreased in the 6b-transgenic plants, we failed to detect any differences in methylation and acetylation of histones in putative promoter regions of the IAA3/SHY2 and IAA6 genes between 6b-transgenic and nontransgenic Arabidopsis plants. It was reported recently that overexpression of TAF-I in HeLa cells induces transcription of genes in chromatin without the acetylation of histone H3 and H4 at the gene that is being transcribed (Kato et al., 2006). Further studies are required to determine how 6b might mediate the transcription of genes without the modification of histones in plant cells.
We have identified two nuclear proteins, in addition to histones, in tobacco that can interact with 6b, namely, Nt SIP1 and Nt SIP2 (Kitakura et al., 2002). It has been reported that the histone chaperones described above are associated with other proteins that are involved in the activation or repression of transcription (Loyola and Almouzni, 2004; Li et al., 2007). The 6b-interacting proteins might play roles in recruiting 6b to specific regions of chromatin, which might be affected by 6b.
Possible Roles of 6b in the Formation of Crown Gall Tumors
Although the 6b gene induces the proliferation of plant cells (see Introduction), the effect of 6b is relatively weak compared with those of genes for the biosynthesis of auxin (iaaM [tms1] and iaaH [tms2]) and cytokinin (ipt [tmr]) in T-DNAs. The 6b gene might play only an auxiliary role in the formation of crown gall because mutations in 6b do not affect the tumorigenicity of T-DNAs to any great extent (Garfinkel et al., 1981; Joos et al., 1983). However, conservation of 6b in the T-DNA regions of many Ti plasmids (Otten and De Ruffray, 1994) suggests a role for the gene in the neoplastic diseases caused by transformation with T-DNA. It has been proposed that the 6b gene might play a more critical role in tumorigenicity in certain specific species of the host plant (Hooykaas et al., 1988). If the efficiency of the formation of crown gall tumors depends on levels of expression of plant cell division–related genes that might be supported by combinatorial functions of the products of the hormone-synthesizing genes from the T-DNA mentioned above and of 6b, which encodes a histone chaperone–like protein, 6b might play a more critical role in the formation of crown galls in host plants that are relatively insensitive to the actions of the phytohormones. By contrast, 6b might be less important in host plants that are sensitive to these phytophormones. The observation that 6b affected the expression of plant genes via its histone chaperone–like activity is consistent with such a hypothesis.
METHODS
Plant Materials and Transformation
Maintenance and transformation of Nicotiana plants (N. tabacum SR1 and N. benthamiana), BY-2 tobacco cultured cells, and Arabidopsis thaliana (Columbia) plants are described elsewhere (Tanaka et al., 2001; Kitakura et al., 2002; Nishihama et al., 2002).
Agrobacterium 6b Genes
We used AK6b and AB6b genes (both encoding 208–amino acid residues) derived from Agrobacterium tumefaciens AKE10 (Wabiko and Minemura, 1996) and Agrobacterium vitis AB4 (Helfer et al., 2003), respectively. Structures of 6b mutants, AK6bΔA and AK6bΔC, were described in the text and depicted in Supplemental Figure 1 online. In the AK6bΔA sequence (the 564-bp sequence), the 63-bp sequence corresponding to the entire acidic region of 6b (amino acid residues 164 to 184) was deleted from the 6b coding sequence (the 627-bp sequence corresponding to 208–amino acid residues plus the termination codon). In the AK6bΔC sequence, the nucleotide sequence corresponding to the C-terminal region from residues 185 to 208 was deleted.
Yeast Two-Hybrid Screening
The screening system was essentially as described previously (Kitakura et al., 2002). We used the LexA-fused AK6bΔA sequence (Kitakura et al., 2002) as bait, in which the LexA coding sequence had been fused to the initiator codon of the AK6bΔA sequence. A cDNA library was synthesized from poly(A)+ RNA that had been isolated from 3-week-old tobacco plants by the standard protocol with a TimeSaver cDNA synthesis kit (Amersham Biosciences).
Construction of Plasmids
Plasmid DNAs were constructed by amplification by PCR and standard cloning techniques. Details of procedures are available upon request. The 35S promoter–fused AK6b gene was described previously (Kitakura et al., 2002). To produce recombinant proteins in Escherichia coli BL21AI (Invitrogen), DNA constructs were cloned into pET28 (Novagen; Kitakura et al., 2002) and pDEST17 plasmids (Invitrogen). To make a DEX-inducible expression system of the 6b gene, His-T7-AK6b DNA was cloned into the pTA7001 vector (Aoyama and Chua, 1997) to yield pTA7001-His-T7-AK6b, which was used for transformation of Arabidopsis.
Partial amino acid sequences for the N-terminal tags and the junction between the tag and 6b and derivatives are as follows: His-AK6b, His-AK6bΔC, and His-AB6b, MGCCHHHHHHSSGLVPRGSHKAYEM; Trx-His-S-AK6bΔA (amino acid sequence for S-tag), PDLGTDDDDKAMADIGSEFELRRQAYEM; His-T7-AK6b, MGCCHHHHHHSSGLVPRGSHMASMTGGQQMGRGSEFELRRQAYEM; His-T7-HMK-AK6b, MGCCHHHHHHSSGLVPRGSHMASMTGGQQMGRGSEFRRASVRRQAYEM. The boldface M indicates the first Met residue in 6b, and underlining indicates the T7 epitope.
Protein Gel Blot Assay
Protein gel blot assays were performed as described by Sambrook et al. (1989). Recombinant histones H2A, H2B, H3, and H4 were purchased from Upstate Biotechnology. To label recombinant 6b protein in vitro, we generated a DNA construct that included the DNA sequence that encoded the amino acid sequence that is specific for phosphorylation by protein kinase A (the HMK site, RRASV; Blaner and Rutter, 1992) and the DNA sequence that encoded the recombinant protein for biosynthesis of the His-T7-HMK-AK6b protein. We radiolabeled His-T7-HMK-AK6b using protein kinase A (Sigma-Aldrich) and [γ-32P]ATP and used the labeled protein as probe in the protein gel blot analysis. Core histones were applied to a Hybond C Extra membrane (Amersham Biosciences), and blotting analysis was performed as described by Sambrook et al. (1989).
BiFC Assay
The BiFC assay with leaves of N. benthamiana was performed basically as described by Abe et al. (2005). GFP-fused HDT1 and GFP-fused AS2 (Ueno et al., 2007) were used as internal controls. For infiltration of Agrobacterium, we used acetosyringone (Wako) at 100 μg/mL in 10 mM MgCl2. Fluorescence due to GFP and YFP in leaf cells was observed with a confocal laser scanning microscope (LSM 510 META; Carl Zeiss).
Isolation of Nuclei
Nuclei were isolated from BY-2 cells as described previously (Masuda et al., 1991). BY-2 cells were washed twice with buffer A (9% [w/v] sorbitol, 25 mM MES-KOH, pH 5.6, and 5 mM CaCl2), suspended in an enzyme mixture (9% [w/v] sorbitol, 3% [w/v] cellulase Onozuka R-10 [Yakult], and 1% [w/v] Macerozyme R10 [Yakult], pH 5.5), and incubated at 25°C for 40 min. After cells had been washed with buffer A, they were suspended in ice-cold buffer B (25 mM MES-KOH, pH 5.6, 5 mM MgCl2, 10 mM KCl, 0.35 M sucrose, 30% [w/w] glycerol, and a protease inhibitor cocktail [Roche]) and then homogenized with a glass homogenizer. The homogenate was filtered successively through a series of stainless-steel screens of decreasing pore diameter (37, 25, and 22 μm), and the screens were washed with buffer C (buffer B containing 4% [w/v] Triton X-100). The nuclei in the final filtrate were collected by centrifugation, and the pellet was resuspended in buffer C. The nuclei were subsequently purified twice by Percoll (Amersham Biosciences) density gradient (48 and 32%) centrifugation. The final preparation of nuclei was suspended in a buffer that contained 50% glycerol, 1 mM DTT, 10 mM KCl, 0.35 M sucrose, 10 mM MgCl2, and 20 mM MES-KOH, pH 6.0, and stored at −80°C prior to use. All manipulations were performed at 4°C.
Preparation of Chromatin from Tobacco BY-2 Cells
The method for the isolation of chromatin has been described previously (Sealy et al., 1989). Isolated nuclei were disrupted in lysis buffer (10 mM PIPES-HCl, pH 6.8, 10 mM EDTA, and the protease inhibitor cocktail), and centrifuged at 6000g for 10 min. The pellet and the supernatant were recovered as the chromatin fraction and the nucleoplasmic fraction, respectively. An aliquot of each fraction was subjected to SDS-PAGE and protein gel blotting analysis using catalase-specific antibodies, histone H3–specific antibodies (Cell Signaling Technology), and T7-specific antibodies (Novagen).
Analysis of Protein–Protein Interactions
The interaction between histone H3 and 6b was examined essentially as described elsewhere (Kitakura et al., 2002), with the exception that the binding buffer was replaced by Tris-buffered saline that contained 10 mM MgCl2 and protease inhibitor cocktail. Isolated nuclei (1 × 106) were resuspended in 500 μL of MNase buffer (20 mM Tris-HCl, pH 7.5, 100 mM KCl, 2 mM MgCl2, 1 mM CaCl2, 0.3 M sucrose, 0.1% [w/v] Triton X-100, 3 units/μL MNase [Amersham Biosciences], and a protease inhibitor cocktail) and incubated at 25°C for 180 min. The reaction was stopped by addition of EDTA and ethyl ethylene glycol-bis (β-aminoethyl ether)-N,N,N‘,N’-tetraacetate (EGTA) to give a final concentration of 5 mM each. Protein complexes were immunoprecipitated with T7-specific antibody-conjugated agarose beads (Novagen), and the agarose beads were then washed three times with Tris-buffered saline. Bound proteins were eluted by boiling in loading buffer for SDS-PAGE and then subjected to SDS-PAGE and protein gel blot analysis with His epitope–specific antibody (Santa Cruz Biotechnology).
Salt Extraction
Nuclei (1 × 106) were incubated in 100 μL of lysis buffer that contained NaCl at various concentrations. After a 30-min incubation on ice, samples were centrifuged at 12,800g for 15 min at 4°C to yield the supernatant and pellet fractions, which were analyzed by protein gel blotting with T7 epitope–specific (Novagen) and histone H3–specific (Cell Signaling Technology) antibodies.
Supercoiling Assay
Recombinant His-T7-6b, His-AB6b, Trx-His-S-6bΔA, and His-6bΔC were produced in E. coli cells and affinity purified on HIS-Select cobalt affinity gel (Sigma-Aldrich). GST-hNAP-1 was purified with glutathione Sepharose (Amersham Biosciences), and then the GST-tag was removed by PreScission protease (Amersham Biosciences). Proteins were subjected to chromatography on Q Sepharose Fast-Flow (Amersham Biosciences). Core histones were purified from HeLa cells as described previously (Simon and Felsenfeld, 1979).
The supercoiling assay was performed as described by Fujii-Nakata et al. (1992). The reaction was performed at 37°C in a buffer (30 μL) that contained 10 mM Tris-HCl, pH 7.5, 100 mM NaCl, 2 mM MgCl2, and 0.1% (w/v) BSA. To obtain relaxed and closed-circular DNA, negatively supercoiled and closed-circular DNA (0.2 μg) of pBR322 was preincubated with 2 units of topoisomerase I (Invitrogen) at 37°C for 30 min in the above-mentioned buffer (10 μL). Core histones, which had been prepared as described above, were also preincubated with recombinant hNAP-1 or recombinant derivatives of 6b (His-T7-AK6b, His-AB6b, Trx-His-S-AK6bΔA, and His-AK6bΔC) at 37°C for 15 min in the same buffer (20 μL). The two reaction mixtures were combined and incubated for 45 min. Each reaction was stopped by addition of 3 μL of stop buffer (2% [w/v] SDS and 0.5 mg/mL proteinase K [Gibco BRL]) and incubation at 37°C for 15 min. After the reaction, plasmid DNA was extracted with phenol and chloroform and precipitated in ethanol. Plasmid DNA was subjected to agarose gel electrophoresis (1% agarose) and visualized by staining with ethidium bromide.
MNase Assay
Relaxed pBR322 plasmid DNA (0.2 μg), core histone (200 ng), and recombinant hNAP-1 and His-T7-AK6b were incubated in 30 μL of assembly buffer for 45 min at 37°C. The mixture was supplemented with 100 mM CaCl2 (final concentration) and MNase (0, 0.5, 1, or 2 units) and then incubated at 37°C for 10 min. The reaction was stopped by addition of stop buffer (see above). Digested DNA was extracted, subjected to polyacrylamide (6%) gel electrophoresis, and visualized by staining with ethidium bromide.
Microarray Analysis
Microarray analysis was performed basically as described by Tsukagoshi et al. (2007). Total RNA was isolated from nontransgenic and the 35S:AK6b-transgenic Arabidopsis plants (13 d after germination). Cy3- and Cy5-labeled cDNA probes were synthesized and hybridized to the Arabidopsis 3 oligonucleotide microarray (Agilent Technologies) according to the manufacturer's instructions. The microarray analysis was performed with two independently isolated RNA samples. Feature extraction and image analysis software (Agilent Technologies) was used to locate and delineate each spot in the array and to integrate each spot's intensity, filtering, and normalization.
Analysis of Levels of IAA Transcripts by the DEX-Inducible System of 6b Expression
Transgenic Arabidopsis (line YU29-24) plants, which carried the DEX-inducible His-T7-AK6b construct, were grown on solidified Murashige and Skoog medium. The plants (14 d after germination) were transferred to liquid Murashige and Skoog medium that contained 10 μM DEX and incubated for 0, 6, 24, and 48 h with rotation at 120 rpm in darkness. The plants were collected, and total proteins and poly(A)+ RNAs were prepared. Levels of transcripts of IAA3/SHY2 and IAA6 were measured by quantitative RT-PCR. Reverse transcription was performed using Ready-To-Go You-Prime First-Strand Beads (Amersham Biosciences) and oligo(dT) primers. The primers used for RT-PCR were as follows: for α-tubulin (TUBA; as a control), pU51 and pU52 (Semiarti et al., 2001); for IAA3/SHY2, IAA3-F (5′-AACATCCCCTCCTCGAAAGG-3′) and IAA3-R (5′-TCTTCTTACTCTGAATGTTGTTCTTCCT-3′); and for IAA6, IAA6-F (5′-GGATGCTCGTCGGAGATGTAC-3′) and IAA6-R (5′-GCATCTGATCTCTTCACGATCCT-3′). Quantitative PCR was performed in the presence of the double-stranded DNA-specific dye SYBR Green (Applied Biosystems). Amplification was monitored in real time with the 7500 real-time PCR system (Applied Biosystems). Amounts of His-T7-AK6b protein and those of His-T7-AK6b transcripts were also measured by protein gel blotting and RT-PCR, respectively.
Chromatin Immunoprecipitation Assay
The procedure for the chromatin immunoprecipitation assay was described previously (Johnson et al., 2002). Thirteen-day-old AK6b-transgenic and nontransgenic Arabidopsis plants were harvested and immersed in buffer A (0.4 M sucrose, 10 mM Tris-HCl, pH 8, 1 mM EDTA, 1 mM PMSF, and 1% [w/v] formaldehyde) under a vacuum for 10 min. Gly was added to a final concentration of 0.1 M, and incubation was continued for an additional 5 min. Plants were then washed and frozen in liquid nitrogen. Approximately 0.3 g of leaves were ground for each immunoprecipitation, and ground leaf tissue was suspended in 1 mL of the lysis buffer (Johnson et al., 2002). All other procedures were performed as described by Johnson et al. (2002). Salmon sperm DNA/protein A agarose product and antibodies (antidimethyl-histone H3 [Lys9] and anti-acetyl-histone H3) were purchased from Upstate Biotechnology. Proteinase K was purchased from Gibco BRL.
We used primer pairs for quantitative PCR that covered sequences of ∼1500 bp, including 5′-upstream regions plus first-exon regions of the IAA3 and IAA6 genes. All PCRs were performed in 50 μL of reaction mixture, starting with incubation for 5 min at 96°C, which was followed by 23 to 36 cycles (depending on the region being amplified) at 94°C (15 s), 60°C (30 s), and 72°C (1 min). Aliquots of the PCR mixture were removed after various numbers of cycles. Products of PCR were resolved by electrophoresis on a 3% agarose gel and detected with ethidium bromide, and then images were captured on a UV transilluminator.
Accession Numbers
DDBJ/GenBank/EMBL accession numbers for tobacco cDNAs that were newly isolated and characterized in this study and Arabidopsis Genome Initiative locus identifiers for the genes described in this article are listed in Supplemental Table 1 online and as follows: tobacco histone H3.1 cDNA, AB331236 (AT5G65360); tobacco histone H3.2 cDNA, AB355993 (AT4G40030); and H2B-histone fold–like protein cDNA, AB355994/EB681212.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Schematic Representation of the Structural Organization of the AK6b Protein and Its Mutant Derivatives and Characterization of Proteins Used in the Callus-Formation Assay and the Protein Binding Assay.
Supplemental Figure 2. Amino Acid Sequences Deduced from Tobacco cDNAs That Were Identified by Yeast Two-Hybrid Screening.
Supplemental Figure 3. Protein 6b Interacts with Histones H3.1 and H3.1 of Arabidopsis in Vitro.
Supplemental Figure 4. Effects of the Synthesis of His-T7-6b on Levels of IAA2/SHY2 and IAA6 Transcripts in Arabidopsis Plants.
Supplemental Table 1. Clones of Tobacco cDNAs That Were Isolated by Yeast Two-Hybrid Screening.
Supplementary Material
Acknowledgments
We thank M. Maeshima (Nagoya University) for providing catalase-specific antibodies. We also thank Y. Yoshioka (Nagoya University) for preparing the cDNA library and K. Matsuoka (Kyushu University, Japan) for information related to the analysis of tobacco genes. This work was supported, in part, by a grant from the Program for Promotion of Basic Research Activities for Innovative Biosciences (Japan) and by Grants-in-Aid for Scientific Research on Priority Areas (10182102, 14036216, and 15028208) from the Ministry of Education, Science, Culture, and Sports (Japan). S.T. was supported by a grant of the 21st Century Center of Excellence program for Division of Biological Science, Nagoya University, and a research fellowship from the Japan Society for the Promotion of Science for Young Scientists.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Yasunori Machida (yas@bio.nagoya-u.ac.jp).
Online version contains Web-only data.
References
- Abe, M., Kobayashi, Y., Yamamoto, S., Daimon, Y., Yamaguchi, A., Ikeda, Y., Ichinoki, H., Notaguchi, M., Goto, K., and Araki, T. (2005). FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309 1052–1056. [DOI] [PubMed] [Google Scholar]
- Adkins, M.W., Howar, S.R., and Tyler, J.K. (2004). Chromatin disassembly mediated by the histone chaperone Asf1 is essential for transcriptional activation of the yeast PHO5 and PHO8 genes. Mol. Cell 14 657–666. [DOI] [PubMed] [Google Scholar]
- Angelov, D., Bondarenko, V.A., Almagro, S., Menoni, H., Mongélard, F., Hans, F., Mietton, F., Studitsky, V.M., Hamiche, A., Dimitrov, S., and Bouvet, P. (2006). Nucleolin is a histone chaperone with FACT-like activity and assists remodeling of nucleosomes. EMBO J. 25 1669–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoyama, T., and Chua, N.-H. (1997). A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J. 11 605–612. [DOI] [PubMed] [Google Scholar]
- Blaner, M.A., and Rutter, W.J. (1992). Interaction cloning: Identification of a helix-loop-helix zipper protein that interacts with c-Fos. Science 256 1014–1018. [DOI] [PubMed] [Google Scholar]
- Canaday, J., Gérad, J.C., Crouzet, P., and Otten, L. (1992). Organization and functional analysis of three T-DNAs from the vitopine Ti plasmid pTiS4. Mol. Gen. Genet. 235 292–303. [DOI] [PubMed] [Google Scholar]
- Clément, B., Pollmann, S., Weiler, E., Urbanczyk-Wochniak, E., and Otten, L. (2006). The Agrobacterium vitis T-6b oncoprotein induces auxin-independent cell expansion in tobacco. Plant J. 45 1017–1027. [DOI] [PubMed] [Google Scholar]
- Dong, A., Liu, Z., Zhu, Y., Yu, F., Li, Z., Cao, K., and Shen, W.-H. (2004). Interacting proteins and differences in nuclear transport reveal specific functions for the NAP1 family proteins in plants. Plant Physiol. 138 1446–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehsan, H., Reichheld, J.P., Durfee, T., and Roe, J.L. (2004). TOUSLED kinase activity oscillates during the cell cycle and interacts with chromatin regulators. Plant Physiol. 134 1488–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippini, F., Rossi, V., Marin, O., Trovato, M., Costantino, P., Downey, P.M., Lo Schiavo, F., and Terzi, M. (1996). A plant oncogene as a phosphatase. Nature 379 499–500. [DOI] [PubMed] [Google Scholar]
- Fujii-Nakata, T., Ishimi, Y., Okuda, A., and Kikuchi, A. (1992). Functional analysis of nucleosome assembly protein, NAP-1. J. Biol. Chem. 267 20980–20986. [PubMed] [Google Scholar]
- Galichet, A., and Gruissem, W. (2006). Developmentally controlled farnesylation modulates AtNAP1;1 function in cell proliferation and cell expansion during Arabidopsis leaf development. Plant Physiol. 142 1412–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gális, I., Simek, P., Van Onckelen, H.A., Kakiuchi, Y., and Wabiko, H. (2002). Resistance of transgenic tobacco seedlings expressing the Agrobacterium tumefaciens C58-6b gene to growth-inhibitory levels of cytokinin is associated with elevated IAA levels and activation of phenylpropanoid metabolism. Plant Cell Physiol. 43 939–950. [DOI] [PubMed] [Google Scholar]
- Garfinkel, D.J., Simpson, R.B., Ream, L.W., White, F.F., Gordon, M.P., and Nester, E.W. (1981). Genetic analysis of crown gall: fine structure map of the T-DNA by site-directed mutagenesis. Cell 27 143–153. [DOI] [PubMed] [Google Scholar]
- Helfer, A., Clément, B., Michler, P., and Otten, L. (2003). The Agrobacterium oncogene AB-6b causes a graft-transmissible enation syndrome in tobacco. Plant Mol. Biol. 52 483–493. [DOI] [PubMed] [Google Scholar]
- Hooykaas, P.J.J., der Dulk-Ras, H., and Schilperoort, R.A. (1988). The Agrobacterium tumefaciens T-DNA gene 6b is an onc gene. Plant Mol. Biol. 11 791–794. [DOI] [PubMed] [Google Scholar]
- Hu, C.D., Chinenov, Y., and Kerppola, T.K. (2002). Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol. Cell 9 789–798. [DOI] [PubMed] [Google Scholar]
- Ishimi, Y., and Kikuchi, A. (1991). Identification and molecular cloning of yeast homolog of nucleosome assembly protein I which facilitates nucleosome assembly in vitro. J. Biol. Chem. 266 7025–7029. [PubMed] [Google Scholar]
- Ishimi, Y., Yasuda, H., Hirosumi, J., Hanaoka, F., and Yamada, M. (1983). A protein which facilitates assembly of nucleosome-like structures in vitro in mammalian cells. J. Biol. Chem. 94 735–744. [DOI] [PubMed] [Google Scholar]
- Johnson, L., Cao, X., and Jacobsen, S.E. (2002). Interplay between two epigenetic marks: DNA methylation and histone H3 lysine 9 methylation. Curr. Biol. 12 1360–1367. [DOI] [PubMed] [Google Scholar]
- Johnson, L., Mollah, S., Garcia, B.A., Muratore, T.L., Shabanowitz, J., Hunt, D.F., and Jacobsen, S.E. (2004). Mass spectrometry analysis of Arabidopsis histone H3 reveals distinct combinations of post-transcriptional modifications. Nucleic Acids Res. 32 6511–6518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joos, H., Inze, D., Caplan, A., Sormann, M., Van Montagu, M., and Schell, J. (1983). Genetic analysis of T-DNA transcripts in nopaline crown galls. Cell 32 1057–1067. [DOI] [PubMed] [Google Scholar]
- Kakiuchi, Y., Gàlis, I., Tamogami, S., and Wabiko, H. (2006). Reduction of polar auxin transport in tobacco by the tumorigenic Agrobacterium tumefaciens AK-6b gene. Planta 223 237–247. [DOI] [PubMed] [Google Scholar]
- Kato, K., Miyaji-Yamaguchi, M., Okuwaki, M., and Nagata, K. (2006). Histone acetylation-independent transcription stimulation by a histone chaperone. Nucleic Acids Res. 35 705–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitakura, S., Fujita, T., Ueno, Y., Terakura, S., Wabiko, H., and Machida, Y. (2002). The protein encoded by oncogene 6b from Agrobacterium tumefaciens interacts with a nuclear protein of tobacco. Plant Cell 14 451–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzuhara, T., and Horikoshi, M. (2004). A nuclear FK506-binding protein is a histone chaperone regulating rDNA silencing. Nat. Struct. Mol. Biol. 11 275–283. [DOI] [PubMed] [Google Scholar]
- Laskey, R.A., Honda, B.M., Mills, A.D., and Finch, J.T. (1978). Nucleosomes are assembled by an acidic protein which binds histones and transfers them to DNA. Nature 275 416–420. [DOI] [PubMed] [Google Scholar]
- Leemans, J., Langenakens, J., De Greve, H., Deblaere, R., Van Montagu, M., and Schell, J. (1982). Broad-host-range cloning vectors derived from the W-plasmid Sa. Gene 19 361–364. [DOI] [PubMed] [Google Scholar]
- Levesque, H., Delepelaire, P., Rouzé, P., Slightom, J., and Tepfer, D. (1988). Common evolutionary origin of the central portions of the Ri TL-DNA of Agrobacterium rhizogenes and the Ti T-DNAs of Agrobacterium tumefaciens. Plant Mol. Biol. 11 731–744. [DOI] [PubMed] [Google Scholar]
- Li, B., Carey, M., and Workman, J.L. (2007). The role of chromatin during transcription. Cell 128 707–719. [DOI] [PubMed] [Google Scholar]
- Loyola, A., and Almouzni, G. (2004). Histone chaperones, a supporting role in the limelight. Biochim. Biophys. Acta 1677 3–11. [DOI] [PubMed] [Google Scholar]
- Luger, K., Mäder, A.W., Richmond, R.K., Sargent, D.F., and Richmond, T.J. (1997). Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 389 251–260. [DOI] [PubMed] [Google Scholar]
- Masuda, K., Takahashi, S., Nomura, K., and Inoue, M. (1991). A simple procedure for the isolation of pure nuclei from carrot embryos in synchronized cultures. Plant Cell Rep. 10 329–333. [DOI] [PubMed] [Google Scholar]
- Morris, R.O. (1986). Genes specifying auxin and cytokinin biosynthesis in phytopathogens. Annu. Rev. Plant Physiol. 37 509–538. [Google Scholar]
- Munakata, T., Adachi, N., Yokoyama, N., Kuzuhara, T., and Horikoshi, M. (2000). A human homologue of yeast anti-silencing factor has histone-chaperone activity. Genes Cells 5 221–233. [DOI] [PubMed] [Google Scholar]
- Nishihama, R., Soyano, T., Ishikawa, M., Araki, S., Tanaka, H., Asada, T., Irie, K., Ito, M., Terada, M., Banno, H., Yamazaki, Y., and Machida, Y. (2002). Expansion of the cell plate in plant cytokinesis requires a kinesin-like protein/MAPKKK complex. Cell 109 87–99. [DOI] [PubMed] [Google Scholar]
- Okuwaki, M., Matsumoto, K., Tsujimoto, M., and Nagata, K. (2001). Function of nucleophosmin/B23, a nucleolar acidic protein, as a histone chaperone. FEBS Lett. 506 272–276. [DOI] [PubMed] [Google Scholar]
- Okuwaki, M., and Nagata, K. (1998). Template-activating factor-I remodels the chromatin structure and stimulates transcription from the chromatin template. J. Biol. Chem. 273 34511–34518. [DOI] [PubMed] [Google Scholar]
- Otten, L., and De Ruffray, P. (1994). Agrobacterium vitis nopaline Ti plasmid pTiAB4: Relationship to other Ti plasmids and T-DNA structure. Mol. Gen. Genet. 245 493–505. [DOI] [PubMed] [Google Scholar]
- Phelps-Durr, T.L., Thomas, J., Vahab, P., and Timmermans, M.C.P. (2005). Maize rough sheath2 and its Arabidopsis orthologue ASYMMETRIC LEAVES1 interact with HIRA, a predicted histone chaperone, to maintain knox gene silencing and determinacy during organogenesis. Plant Cell 17 2886–2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray-Gallet, D., Quivy, J.P., Scamps, C., Martini, E.M., Lipinski, M., and Almouzni, G. (2002). HIRA is critical for a nucleosome assembly pathway independent of DNA synthesis. Mol. Cell 9 1091–1100. [DOI] [PubMed] [Google Scholar]
- Ream, L.W., Gordon, M.P., and Nester, E.W. (1983). Multiple mutations in the T region of the Agrobacterium tumefaciens tumor-inducing plasmid. Proc. Natl. Acad. Sci. USA 80 1660–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Schwabish, M.A., and Struhl, K. (2006). Asf1 mediates histone eviction and deposition during elongation by RNA polymerase II. Mol. Cell 22 415–422. [DOI] [PubMed] [Google Scholar]
- Sealy, L., Burgess, R.R., Cotten, M., and Chalkley, R. (1989). Purification of Xenopus egg nucleoplasmin and its use in chromatin assembly in vitro. Methods Enzymol. 170 612–630. [DOI] [PubMed] [Google Scholar]
- Semiarti, E., Ueno, Y., Tsukaya, H., Iwakawa, H., Machida, C., and Machida, Y. (2001). The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana regulates formation of a symmetric lamina, establishment of venation and repression of meristem-related homeobox genes in leaves. Development 128 1771–1783. [DOI] [PubMed] [Google Scholar]
- Silljé, H.H., and Nigg, E.A. (2001). Identification of human Asf1 chromatin assembly factors as substrates of Tousled-like kinases. Curr. Biol. 11 1068–1073. [DOI] [PubMed] [Google Scholar]
- Simon, R.H., and Felsenfeld, G. (1979). A new procedure for purifying histone pairs H2A + H2B and H3 + H4 from chromatin using hydroxylapatite. Nucleic Acids Res. 6 689–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spanier, K., Schell, J., and Schreier, P.H. (1989). A functional analysis of T-DNA gene 6b: The fine tuning of cytokinin effects on shoot development. Mol. Gen. Genet. 219 209–216. [DOI] [PubMed] [Google Scholar]
- Spena, A., Schmülling, T., Koncz, C., and Schell, J.S. (1987). Independent and synergistic activity of rol A, B and C loci in stimulating abnormal growth in plants. EMBO J. 6 3891–3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stieger, P.A., Meyer, A.D., Kathmann, P., Fründt, C., Niederhauser, I., Barone, M., and Kuhlemeier, C. (2004). The orf13 T-DNA gene of Agrobacterium rhizogenes confers meristematic competence to differentiated cells. Plant Physiol. 135 1798–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka, H., Onouchi, H., Kondo, M., Hara-Nishimura, I., Nishimura, M., Machida, C., and Machida, Y. (2001). A subtilisin-like serine protease is required for epidermal surface formation in Arabidopsis embryos and juvenile plants. Development 128 4681–4689. [DOI] [PubMed] [Google Scholar]
- Terakura, S., Kitakura, S., Ishikawa, M., Ueno, Y., Fujita, T., Machida, C., Wabiko, H., and Machida, Y. (2006). Oncogene 6b from Agrobacterium tumefaciens induces abaxial cell division at late stages of leaf development and modifies vascular development in petioles. Plant Cell Physiol. 47 664–672. [DOI] [PubMed] [Google Scholar]
- Tinland, B., Fournier, P., Heckel, T., and Otten, L. (1992). Expression of a chimaeric heat-shock-inducible Agrobacterium 6b oncogene in Nicotiana rustica. Plant Mol. Biol. 18 921–930. [DOI] [PubMed] [Google Scholar]
- Tinland, B., Rohfritsch, O., Michler, P., and Otten, L. (1990). Agrobacterium tumefaciens T-DNA gene 6b stimulates rol-induced root formation, permits growth at high auxin concentrations and increases root size. Mol. Gen. Genet. 223 1–10. [DOI] [PubMed] [Google Scholar]
- Tsukagoshi, H., Morikami, A., and Nakamura, K. (2007). Two B3 domain transcriptional repressors prevent sugar-inducible expression of seed maturation genes in Arabidopsis seedlings. Proc. Natl. Acad. Sci. USA 104 2543–2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler, J.K., Adams, C.R., Chen, S.-R., Kobayashi, R., Kamakaka, R.T., and Kadonaga, J.T. (1999). The RCAF complex mediates chromatin assembly during DNA replication and repair. Nature 402 555–560. [DOI] [PubMed] [Google Scholar]
- Ueno, Y., Ishikawa, T., Watanabe, K., Terakura, S., Iwakawa, H., Okada, K., Machida, C., and Machida, Y. (2007). Histone deacetylases and ASYMMETRIC LEAVES2 are involved in the establishment of polarity in leaves of Arabidopsis. Plant Cell 19 445–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wabiko, H., and Minemura, M. (1996). Exogenous phytohormone-independent growth and regeneration of tobacco plants transgenic for the 6b gene of Agrobacterium tumefaciens AKE10. Plant Physiol. 112 939–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterborg, J.H., and Robertson, A.J. (1996). Common features of analogous replacement of histone H3 genes in animals and plants. J. Mol. Evol. 43 194–206. [DOI] [PubMed] [Google Scholar]
- Willmitzer, L., Dhaese, P., Schreier, P.H., Schmalenbach, W., Van Montagu, M., and Schell, J. (1983). Size, location and polarity of T-DNA-encoded transcripts in nopaline crown gall tumors; common transcripts in octopine and nopaline tumors. Cell 32 1045–1056. [DOI] [PubMed] [Google Scholar]
- Workman, J.L. (2006). Nucleosome displacement in transcription. Genes Dev. 20 2009–2017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.