Abstract
The Arabidopsis (Arabidopsis thaliana) AAA ATPase CDC48A was fused to cerulean fluorescent protein and yellow fluorescent protein. AAA ATPases like CDC48 are only active in hexameric form. Förster resonance energy transfer-based fluorescence lifetime imaging microscopy using CDC48A-cerulean fluorescent protein and CDC48A-yellow fluorescent protein showed interaction between two adjacent protomers, demonstrating homo-oligomerization occurs in living plant cells. Interaction between CDC48A and the SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASE1 (SERK1) transmembrane receptor occurs in very restricted domains at the plasma membrane. In these domains the predominant form of the fluorescently tagged CDC48A protein is a hexamer, suggesting that SERK1 is associated with the active form of CDC48A in vivo. SERK1 trans-phosphorylates CDC48A on Ser-41. Förster resonance energy transfer-fluorescence lifetime imaging microscopy was used to show that in vivo the C-terminal domains of CDC48A stay in close proximity. Employing fluorescence correlation spectroscopy, it was shown that CDC48A hexamers are part of larger complexes.
The Arabidopsis (Arabidopsis thaliana) cell division cycle protein CDC48A was previously shown to interact with SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASE1 (SERK1) (Rienties et al., 2005) and to coimmunoprecipitate with SERK1 in Arabidopsis cultured cells and seedlings (Karlova et al., 2006). In living cells the CDC48A protein colocalizes with SERK1 at peripheral endoplasmic reticulum (ER)-based membranes and the plasma membrane (PM). Förster resonance energy transfer (FRET)-fluorescence lifetime imaging microscopy (FLIM) showed that CDC48A interacts with SERK1 at the PM (Aker et al., 2006).
CDC48A is a member of the family of AAA ATPases (ATPases associated with various cellular activities), shown to have various functions in cell division, membrane fusions, and in proteasome- and ER-associated degradation (ERAD) of proteins (Woodman, 2003). The role of AAA ATPases is to generate mechanical force to disrupt or fuse molecular structures by means of ATP binding and hydrolysis. The AtCDC48A protein was shown to play a role in ERAD and in membrane fusions (Rancour et al., 2002; Müller et al., 2005). AAA proteins are present as stacked hexameric rings that are stabilized by the binding of ATP. They are only reported to be active in hexameric form. This study aims to determine the predominant form of the fluorescently tagged CDC48A protein in Arabidopsis protoplasts at the peripheral ER and the PM domains where interactions between CDC48A and the SERK1 receptor take place, and, therefore, to determine if SERK1 is associated with the active form of CDC48A.
CDC48A is 77% identical to the mammalian homolog vasolin-containing protein (VCP) or p97. Recently, conformational and dynamic changes during the ATP hydrolysis cycle of p97 have been studied by cryo-electron microscopy, crystallography, and small-angle x-ray scattering (for review, see Pye et al., 2006). The monomeric protein contains an N-terminal domain, two AAA domains, and a C-terminal domain (Figs. 1 and 3). The N domain is important for binding of adaptor proteins and substrates. The two AAA domains D1 and D2 are responsible for binding and hydrolysis of ATP. The D1 and D2 domains form stacked hexameric rings above each other that are connected by a linker. The ATP-binding site in the D1 domain and the D1-D2 linker are responsible for hexamerization, while the ATP-binding site in the D2 domain possesses the major ATPase activity (Wang et al., 2003a). The N domain is flexibly attached to the D1 domain, and projects out of the ring, facilitating binding to other proteins. During ATP hydrolysis, large conformational changes are transmitted from the D2 domain via the D1 domain onto the N domains, due to binding, hydrolysis, and release of the nucleotide (DeLaBarre and Brunger, 2003, 2005). In addition, the pores within the D1 and D2 rings narrow and widen in different nucleotide states. This causes the rings to rotate relative to each other (Davies et al., 2005). The C-terminal tail was not ordered within the crystal structure, and it is therefore unknown whether it stays close to the D2 ring or projects from the core structure toward the N domain.
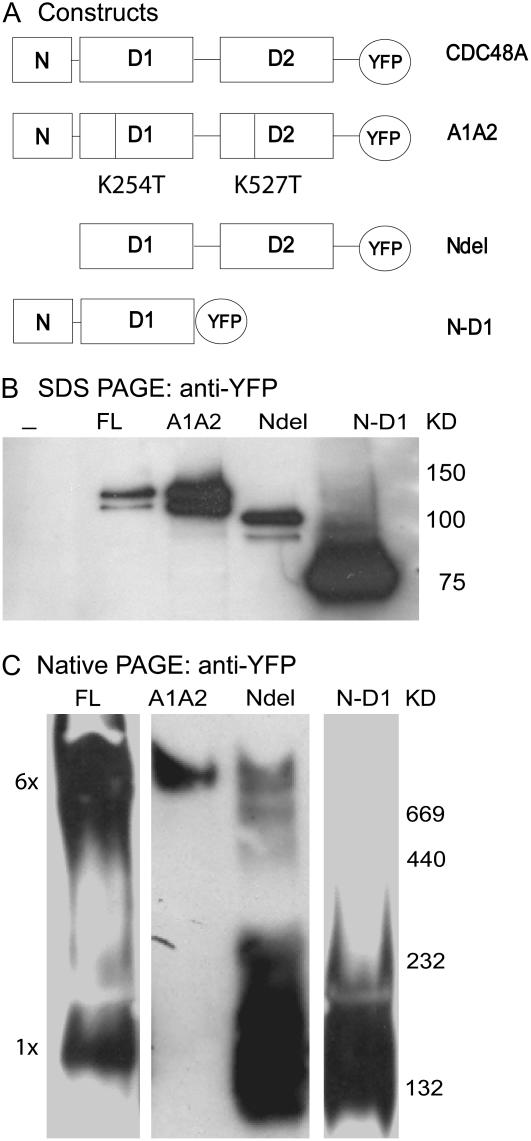
Figure 1.
Expression in Arabidopsis protoplasts of CDC48A, CDC48AA1A2, CDC48ANdel, and CDC48AN-D1 mutants all tagged to YFP. YFP is replaced by CrFP. In A the various mutants are shown schematically. Lysates of transfected cells were submitted to SDS-PAGE in B, to native PAGE in C, and probed with anti-YFP anti-serum. The expected sizes of the denatured proteins are 125 kD for full-length (FL) CDC48A- and CDC48AA1A2-YFP, 96 kD for CDC48ANdel-YFP, and 77 kD for CDC48AN-D1-YFP. Nondenatured hexameric CDC48A-YFP is expected to be between 650 and 750 kD depending on the number of YFP tags.
Figure 3.
Models of the expected hexamers of CDC48A after coexpression of two different protomers. A, Monomeric CDC48A. B, CDC48A fused with CrFP and with YFP (both at the C terminus) are combined and depicted as a hexamer. For simplicity only four fluorophores are drawn at adjacent positions at the C terminus, but in fact these can be randomly positioned at each C domain. C, YFP fused to the N terminus and CrFP to the C terminus. D, YFP and CrFP fused to the N terminus. E and F, CDC48AN-D1 and CDC48ANdel, each depicted as a monomer.
For Arabidopsis CDC48A the crystal structure is not known, but in vitro-produced proteins form a hexamer while PUX1 (plant UBX domain-containing protein) facilitates CDC48A oligomer disassembly (Rancour et al., 2004). Up to now hexamerization of AAA ATPases was only shown in vitro or in total cell lysates, studies that do not reveal any spatial information on the oligomerization status of proteins in a living cell. Therefore, the protein was fused to the GFP variant monomeric cerulean fluorescent protein (CrFP) or to yellow fluorescent protein (YFP), and the hexamerization of CDC48A proteins in living cells was monitored by FRET-FLIM. Employing this technique, oligomerization of the CDC48A proteins in living cells was shown to be nonuniform. In addition, intersubunit distances in the oligomeric CDC48A protein could be calculated, revealing that the C-terminal domains stayed in close proximity rather than protruding out of the molecule.
It was also shown that SERK1 interacts with the N terminus as well as the C terminus of CDC48A (Aker et al., 2006). After performing a trans-phosphorylation reaction with the SERK1 kinase domain, only Ser-41, a Ser residue found to be in the N domain of CDC48A, was phosphorylated.
To investigate the diffusion of the CDC48A protein in cells, fluorescence correlation spectroscopy (FCS), in which fluorescence intensity fluctuations caused by diffusion of fluorescent molecules in and out of a femtoliter volume are monitored in time, was employed. These fluctuations give information about diffusion times of proteins through the volume and, hence, about the size of protein complexes.
Our results show that the oligomeric form of CDC48A in living cells is primarily hexameric and that the fluorescently tagged CDC48A is still able to form hexamers. The SERK1 receptor interacts with CDC48A at the same locations where oligomerization of the CDC48A protein is shown. We conclude therefore that SERK1 interacts with the hexameric form of CDC48A. Using FCS the presence of CDC48A in larger protein complexes in vivo was predicted.
RESULTS
Expression and Localization of CDC48A Wild-Type and Mutant Proteins
The CDC48A protein and various mutants used in this study are depicted in Figure 1A. CDC48A consists of the N domain, the D1 and D2 domains connected by a linker, and the C-terminal tail. The YFP tag is shown here as a C-terminal fusion, and in appropriate constructs it was replaced by CrFP and/or placed at the N terminus. The CDC48AA1A2 mutant contains the K254T and K527T mutations in the Walker A motifs of the D1 and D2 domains, which inhibit ATP binding and in vitro severely delay hexamerization in the mammalian homolog p97/VCP (Wang et al., 2003a). The N-deletion (CDC48ANdel) mutant lacks the N domain (amino acids 1–188), which in other homologs is responsible for binding to cofactors like P47, Ufd1-Npl4, and ubiquitinated substrates (Meyer et al., 2000; Dreveny et al., 2004). The CDC48AN-D1 mutant lacks the D2 domain and the linker region between D1 and D2 (from amino acid 448 to the C terminus). The D1-D2 linker was shown to be important for hexamerization of VCP (Wang et al., 2003b) in vitro.
To investigate whether the CDC48A-YFP proteins are indeed capable of forming a hexameric complex, the CDC48A-YFP and mutant genes were transiently expressed in Arabidopsis protoplasts and analyzed on denaturing and nondenaturing PAGE. Western-blot analysis (Fig. 1B) shows that in protoplasts the various proteins confirm the expected sizes of approximately 125 kD for CDC48A and CDC48AA1A2, 96 kD for CDC48ANdel 1-188, and 77 kD for CDC48AN-D1. The estimated size of the full-length CDC48A protein after native PAGE is about 700 to 750 kD, in line with the calculated size of 625 to 750 kD, the difference depending on the number of GFP tags present (Fig. 1C). However, about 5% to 10% of the YFP-tagged protein appears to remain as monomers. Surprisingly, the CDC48AA1A2 protein is also predominantly found as a hexamer on native PAGE, despite the mutations delaying hexamerization. In a longer exposure of the western blot, CDC48AA1A2 does show a monomeric protein fraction (data not shown). CDC48ANdel 1-188 shows bands of various sizes, but most prominently the monomeric band. In a study by Wang and coworkers, a VCP mutant lacking amino acids 1 to 141 of the N domain is still able to hexamerize in vitro (Wang et al., 2003b). Our data show that the CDC48ANdel protein lacking the complete N domain (amino acids 1–188) is most likely in the monomeric form. The CDC48AN-D1 protein runs as a monomer, in line with the results presented by Wang et al. (2003b).
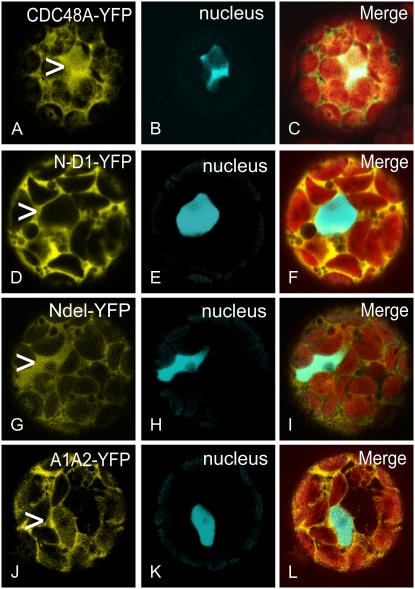
To compare the localizations of the different CDC48A mutant proteins, they were coexpressed with CDC48C-CrFP, shown to localize solely to the nucleus (Aker et al., 2006). The localization of the CDC48A protein is at the PM, in the cytoplasm, and in the ER, but a small fraction also was observed in the nucleus (Aker et al., 2006). In Figure 2, images of the nucleus of CDC48A are shown in the focal plane. CDC48A colocalizes with CDC48C in the nucleus (Fig. 2, A–C) and also localizes to the PM and the cytoplasm. The PM is not in the focal plane and can therefore not be seen. The merge shows an overlay of the cells containing chloroplasts in red, CDC48A in yellow, and the nucleus in cyan. The CDC48AN-D1 protein (Fig. 2, D–F) is found in the cytoplasm but not in the nucleus (arrowhead in Fig. 2D). This can be explained by the Tyr-808 residue, comparable to Tyr-834 in yeast cdc48p and shown to be required for nuclear translocation (Madeo et al., 1998) and that is present in the deleted part of CDC48A. Apparently, although the two nuclear localization signals are still present in the mutant, these are not sufficient for nuclear targeting. A similar observation was made for the mammalian CDC48 ortholog VCP (Indig et al., 2003). To our surprise, the CDC48ANdel protein (Fig. 2, G–I), lacking the nuclear localization signal sequences, still localizes to the nucleus (Fig. 2, compare G with H). For VCP, cytoplasmic localization was described when the N domain of the protein was removed (Indig et al., 2003), and Wang et al. (2003b) found that hexamerization of this protein in vitro could still take place. The CDC48AA1A2 protein (Fig. 2, J–L) is found in the cytoplasm as well as in the nucleus. We conclude based on the hexamerization status in vitro and the localization of the mutant proteins in cells that complete hexamerization of CDC48A is not required for nuclear localization.
Figure 2.
Localizations of CDC48A- or CDC48AN-D1- and CDC48ANdel-YFP. CDC48C-CrFP was used as a nuclear marker (B, E, H, and K). Images were taken at the plane of the nucleus and the merged picture shows the combination of both images together with the chloroplasts in red (C, F, I, and L). CDC48A-YFP localizes to the cytoplasm and the nucleus (compare position of arrowhead in A with image B). The localization at the PM is not visible in this focal plane. CDC48AN-D1 does localize to the cytoplasm but not to the nucleus (compare the arrowhead in image D with E). CDC48ANdel does localize to the cytoplasm and to the nucleus (compare the arrowhead in G with H). The localization of CDC48AA1A2 (J) is comparable with that of CDC48A (A).
Oligomerization of CDC48A at Locations with SERK1 Interaction
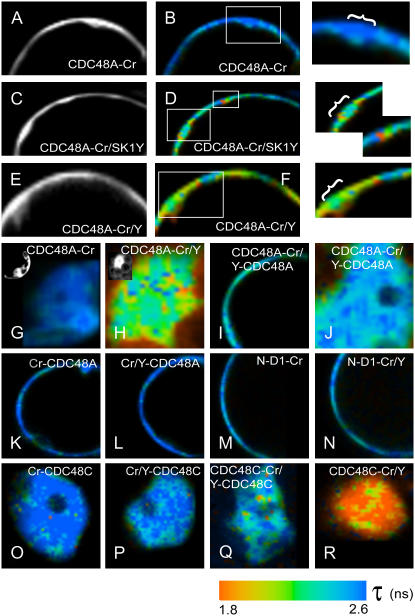
To investigate if the SERK1 receptor interacts in vivo with the hexameric or the monomeric form of CDC48A, the oligomerization of CrFP- and YFP-fused CDC48A protomers in vivo was studied. The CDC48A protein was fused at the C terminus with CrFP or YFP and expressed in Arabidopsis protoplasts. For the FLIM measurements, CDC48A protomers C-terminally fused to CrFP and YFP (Fig. 3A) were coexpressed, hypothetically leading to a hexamer as shown in the model in Figure 3B. Only four of the six fluorophores are drawn at adjacent positions at the C terminus, but in fact these can be randomly positioned at each C domain. The other combinations used are depicted in Figure 3, C to F. Figure 3C shows a hexamer with the donor at the C terminus and the acceptor at the N terminus; Figure 3D shows a hexamer of CDC48A tagged to CrFP and YFP (both at the N terminus); and Figure 3, E and F, shows a monomer of CDC48AN-D1 and CDC48ANdel with tags only at the C terminus.
A maximum energy transfer efficiency of around 50% can only be achieved when fluorophores are approximately 5 nm apart. The distance between two adjacent C termini is not known, but the inner pore diameter of the hexamer is 5.7 nm based on the crystal structure of p97/VCP (Zhang et al., 2000; DeLaBarre and Brunger, 2003; Huyton, 2003). Interaction not only between two adjacent fluorophores but also between two fluorophores on either side of the pore should therefore theoretically be possible. To measure significant energy transfer, the amount of labeled protein has to be relatively high as compared to the amount of endogenous CDC48A. Based on the previous observation that in transfected protoplasts the fluorescent CDC48A level is around 8 times higher than the endogenous level (Aker et al., 2006), we assume that in a hexamer most of the CDC48A protomers contain a fluorescent tag.
First, we investigated if at the locations where CDC48A-SERK1 interaction takes place also oligomerization of the CDC48A proteins occurs. In Figure 4A, the intensity image of the donor molecule CDC48A-CrFP is shown. In the false-color image (Fig. 4B), the average lifetime τ at the PM is shown to be approximately 2.5 to 2.6 ns. As reported previously, coexpression of CDC48A-CrFP and SERK1-YFP resulted in a reduction of the fluorescence lifetime in a punctuate pattern at the membrane (Fig. 4, C and D, inserts). Part of the areas where SERK1 and CDC48A interacted, colocalized with an ER marker but not with an endocytic marker (Aker et al., 2006). After coexpression of CDC48A-CrFP and CDC48A-YFP, interaction was found in a comparable but more widespread area at the PM (Fig. 4, F, insert). We therefore propose that in those membrane areas where interaction between SERK1 and CDC48A occurs, a substantial amount of CDC48A is in oligomeric form. In Table I, the results of all the measurements are listed. In the top portion, the average lifetimes of the full-length CDC48A proteins are listed, in the middle portion the lifetimes of the monomeric mutant CDC48AN-D1, and in the lower portion the lifetimes of CDC48C. The left side refers to the measurements at the PM; the right side refers to the measurements in the nucleus. The unquenched lifetime of the donor CDC48A-CrFP protein is 2.5 ns in the nucleus as well as at the PM. Figure 4G shows a false-color image of the nucleus of a protoplast expressing only the donor molecule CDC48A-CrFP. In the left upper corner, the corresponding intensity image is shown.
Figure 4.
Interactions between CDC48A-tagged protomers based on FRET measured by FLIM. A, Intensity images of the PM of part of a protoplast expressing the donor molecule CDC48-CrFP alone; B, the false-color or lifetime image. A long lifetime, giving a dark blue color, means no interaction; a reduction in donor lifetime, generating a shift toward orange, means interaction. The combination of CDC48A-CrFP and SERK1-YFP shows a reduction in lifetime at specific regions at the PM (C and D plus inserts). A combination of CDC48A-CrFP and CDC48A-YFP proteins shows a reduction in lifetime at the PM in the same regions where SERK1 and CDC48A interact (F plus insert) and in the nucleus (H). In the left top corners, intensity images are shown. A combination of CDC48A-CrFP and YFP-CDC48A at the PM is shown in I and in the nucleus in J. K and L, Images of CrFP-CDC48A alone at the PM (K) and in combination with YFP-CDC48A (L), showing no reduction in lifetime. M and N, Lifetime images of the CDC48AN-D1-CrFP protein as the donor (M) and the combination with a CDC48AN-D1-YFP protein (N). O and P, Lifetime images of the CrFP-CDC48C donor alone (O) and in combination with YFP-CDC48C (P). Q and R, Combination of CDC48C-CrFP with YFP-CDC48C (Q) or with CDC48C-YFP (R) shows a reduction in lifetime.
Table I.
FRET-FLIM analysis of CDC48A, ND1 mutant, and CDC48C, fused to CrFP or YFP
d, Diameter; Eff., efficiency. Lifetime τ is determined as described in “Materials and Methods.” The distance d (nm) and the FRET efficiency between the CrFP and YFP fluorophores are calculated via Equations 1 and 2. N-D1 proteins are not localized in the nucleus, CDC48C proteins not at the PM. A-Cr is CDC48A-CrFP; A-Cr/Y-A is CDC48A-CrFP/YFP-CDC48A; Cr-A/Y-A is CrFP-CDC48A/YFP-CDC48A; A-Cr/A-Y is CDC48A-CrFP/CDC48A-YFP; N-D1-Cr is N-D1-CrFP; N-D1-Cr/Y is N-D1-CrFP/N-D1-YFP; Cr-C is CrFP-CDC48C; C-Cr/Y-C is CDC48C-CrFP/YFP-CDC48C; Cr-C/Y-C is CrFP-CDC48C/YFP-CDC48C; and C-Cr/C-Y is CDC48C-CrFP/CDC48C-YFP.
| Protein | τ PM | d | FRET Eff. | τ Nucleus | d | FRET Eff. |
|---|---|---|---|---|---|---|
| ns | nm | % | ns | nm | % | |
| A-Cr | 2.52 ± 0.03 | 2.53 ± 0.04 | ||||
| A-Cr/Y-A | 2.34 ± 0.04 | 7.6 | 7.2 | 2.44 ± 0.03 | 8.5 | 5.6 |
| Cr-A/Y-A | 2.47 ± 0.06 | 9.5 | 1.9 | 2.55 ± 0.06 | 10.6 | 0 |
| A-Cr/A-Y | 2.18 ± 0.05 | 6.8 | 13.5 | 2.20 ± 0.07 | 6.9 | 12.7 |
| N-D1-Cr | 2.48 ± 0.06 | |||||
| N-D1-Cr/Y | 2.40 ± 0.03 | 8.7 | 3.2 | |||
| Cr-C | 2.59 ± 0.03 | |||||
| C-Cr/Y-C | 2.43 ± 0.04 | 7.8 | 6.4 | |||
| Cr-C/Y-C | 2.44 ± 0.10 | 7.9 | 5.8 | |||
| C-Cr/C-Y | 2.09 ± 0.05 | 6.4 | 19.3 |
Subsequently, CDC48A-CrFP- and CDC48A-YFP-tagged proteins were coexpressed. In Figure 4H, an example of a false-color image is shown for the nucleus. This combination shows a significant reduction in lifetime at the PM (Fig. 4F: 2.18 ± 0.04 ns on average; Table I) as well as in the nucleus (2.20 ± 0.07 ns). From the differences in lifetime between donor alone and donor plus acceptor, the distance between the fluorophores can be calculated. The average distance between two fluorophores both at the C terminus is 6.8 nm, with a FRET efficiency of 13.5%.
FLIM as a Tool to Measure Distances between Subunits in Oligomeric Complexes
The distances between the N domains and C domains in the CDC48A protein, as calculated from the crystal structure of the mammalian homolog VCP/p97 (DeLaBarre and Brunger, 2003), are approximately 8 nm, which is just outside the range at which FRET can be detected between two fluorophores. To determine this distance for the CDC48A protein, a combination of CDC48A-CrFP and YFP-CDC48A was expressed (see Fig. 3C). The mixed protomers showed a minor reduction in the fluorescence lifetime (A-Cr/Y-A, τ = 2.34 ± 0.04 ns; Fig. 4I; Table I) at the PM and 2.44 ± 0.03 ns in the nucleus (A-Cr/Y-A; Fig. 4J; Table I). Based on the FRET efficiencies between the CrFP and YFP fluorophores, the estimated distance between the N terminus and C terminus using Equations 1 and 2 is between 7.6 nm (for the PM) and 8.5 nm (for the nucleus), which is in agreement with the estimated value for p97/VCP of 8 nm. The crystal structure of p97/VCP also showed that, at the plane of the N domains, the hexameric protein has a diameter of approximately 15 nm. Therefore, fluorescent molecules at opposite positions in CDC48 cannot show FRET, so any interaction resulting in a reduction in lifetime would have to originate from adjacent protomers. CrFP- and YFP-CDC48A proteins were coexpressed (Fig. 4L) and the fluorescence lifetimes were compared with the donor (Fig. 4K). No significant reduction in lifetime was measured at the PM (Cr-A/Y-A, τ = 2.47 ± 0.06 ns; Table I) or in the nucleus (Cr-A/Y-A, τ = 2.55 ± 0.06 ns; Table I). The distance between the N domains is calculated to be between 9.5 nm (for the PM) and 10.6 nm (for the nucleus), again confirming the structure as determined for p97/VCP. The FRET efficiency is only 1.9%, meaning that there is no interaction at all.
The CDC48AN-D1-CrFP/-YFP combination did not show a reduction in the fluorescence lifetime (N-D1Cr/Y, τ = 2.40 ± 0.03 ns; Fig. 4N; Table I) compared to the donor alone (N-D1-Cr, τ = 2.48 ± 0.06 ns; Fig. 4M; Table I).
The FRET efficiency was only 3.2%, confirming the monomeric nature of this mutant (Fig. 1C) and demonstrating the importance of the D1-D2 linker and/or the D2 domain in the oligomerization.
In general, the reduction in lifetime between the donor group CrFP alone and the donor plus the acceptor group YFP was less in the nucleus than in the PM. The reason for differences in lifetimes at the PM or in the nucleus of the same protein could be due to the difference in refractive indices of the environment (Borst et al., 2005).
To confirm the relevance of our protein-protein distance measurements using FRET-FLIM, we next investigated the characteristics of CDC48C, another member of the Arabidopsis ATPase family that, in contrast with CDC48A, is only present in the nucleus (Aker et al., 2006). Based on the presence of Walker A and B motifs, CDC48C is classified as an AAA ATPase as well and, therefore, the same hexameric structure is expected. The average donor lifetime of CrFP-CDC48C was 2.59 ± 0.03 (Cr-C; Fig. 4K; Table I). The combination of CrFP- and YFP-CDC48C showed a lifetime of 2.44 ± 0.1 ns (Cr-C/Y-C; Fig. 4P; Table I), a significant reduction compared to the donor alone. The FRET efficiency was 5.8%. The combination of CDC48C-CrFP and YFP-CDC48C protomers also showed a minor but significant reduction in lifetime (C-Cr/Y-C, τ = 2.43 ± 0.04 ns; Fig. 4Q; Table I) comparable with the same combination for CDC48A. Coexpressed CDC48C-CrFP and -YFP proteins showed a large reduction in donor lifetime (C-Cr/C-Y, τ = 2.09 ± 0.05 ns; Fig. 4R; Table I) comparable to that of the CDC48A proteins. The distance between the fluorophores was 6.4 nm, with a FRET efficiency of 19.3%.
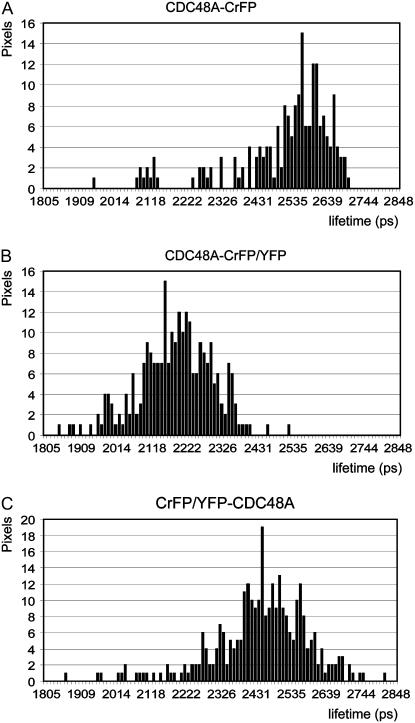
When analyzing FRET-FLIM measurements, average lifetimes are calculated from lifetime distributions in order to compare different combinations. However, spatial distributions may be more illustrative for changes in interactions (Tonaco et al., 2005). In Figure 5, examples of lifetime distribution histograms at the PM are shown for CDC48A-CrFP alone (Fig. 5A), CDC48A-CrFP in combination with CDC48A-YFP (Fig. 5B), and a combination of CrFP-CDC48A and YFP (Fig. 5C). These histograms are examples recorded from protoplasts displaying fluorescence lifetimes as in images Figure 4, B, F, and L, respectively. The nucleus in image Figure 4H is surrounded by chlorophyll, which has a very short lifetime (normally shorter than 1,500 ps) and is indicated in a red color. These lifetimes were not taken into account in the histograms shown in Figure 5 and for the calculations of the average lifetimes. The lifetime distribution histograms clearly show the shift to lower lifetime values when CDC48A-YFP and CDC48A-CrFP were coexpressed (Fig. 5B), while the lack of interaction between the N-terminally placed fluorescent groups is also clearly reflected in the absence of a shift to lower lifetime values (Fig. 5C). For comparison of the various combinations of proteins, combined distribution histograms of five representative experiments for each protein combination were plotted (see supplemental data).
Figure 5.
Representative lifetime distribution histograms measured with FLIM at the PM of Arabidopsis protoplasts. A, Histogram of the CDC48A donor alone. B, Histogram of a mixture of CDC48A tagged to CrFP and YFP at the C terminus, which shifted to lower lifetime values due to interaction. C, Histogram of a mixture of CDC48A tagged to CrFP and YFP at the N terminus. No significant shift to lower lifetime values is visible.
Taken together, our observations show YFP/CrFP proteins fused at the C terminus of CDC48A are within the maximal distance for measuring FRET and do interact, which is only possible when the protomers homo-oligomerize and the C termini stay close together rather than protruding out of the molecule. This oligomerization takes place in the ER membrane-based regions at the PM, where SERK1-CDC48A interaction takes place. A combination of CrFP- and YFP-CDC48A proteins shows no reduction in lifetime, suggesting that the N domains of CDC48A, in line with the mushroom-like shape structure of p97/VCP, are not within FRET distance and cannot interact. The N termini of CDC48C, in contrast with CDC48A, are within FRET distance and do interact, suggesting that the N-terminal domains of CDC48C are differently organized and probably closer to each other.
SERK1 Phosphorylates Ser-41 in the N Domain of CDC48A
In previous work it was shown that SERK1 interacts both with the N domain as well as with the C terminus of CDC48A. To investigate if this interaction involves trans-phosporylation of CDC48A by the SERK1 kinase, an in vitro kinase experiment was performed. The SERK1 kinase domain and full-length CDC48A were produced in Escherichia coli as fusions to glutathione S-transferase (GST), purified and incubated with [γ32-P]ATP. After incubation, the mixture was analyzed by SDS-PAGE and labeled proteins were detected with a PhosphorImager. The SERK1 kinase domain is heavily autophosphorylated (Fig. 6A, lanes 1, 2, and 3) but also trans-phosphorylates CDC48A (arrowhead, lane 1). Next, the experiment was repeated with cold ATP. The 125-kD CDC48A proteins were isolated from the gel, digested with trypsin, and the resulting peptides analyzed by liquid chromatography (LC)-tandem mass spectrometry (MS/MS). Only in the sample containing the SERK1 kinase, one phosphorylated Ser residue was found at position 41 of CDC48A, which is located in the N domain. Unfortunately, we did not have any peptide coverage in the C-terminal tail (from amino acid 775 onward; Fig. 6B), where three Sers, three Thrs, and one Tyr are present, so there may be additional targets for SERK1 phosphorylation.
Figure 6.
In vitro trans-phosphorylation of CDC48A by SERK1 kinase protein. A, Left, Phosphor image of CDC48A trans-phosphorylated by the SERK1 kinase domain. GST-CDC48A, GST alone, or water was incubated with the SERK1 kinase domain (SK1-KD) in a buffer containing [γ32-P]ATP (lane 1, 2, and 3, respectively). Two control experiments were performed without SK1-KD. SK1-KD is autophosphorylated and trans-phosphorylates CDC48A (arrowhead, lane 1). A, Right, Equal loading and size control of GST-CDC48A, GST, and SK1-KD protein. B, Peptide coverage of CDC48A in two reactions containing CDC48A alone or CDC48A plus SK1-KD was 66% and 68%, respectively (colored in gray). Ser-41 of CDC48A was phosphorylated by SK1-KD.
The CDC48A Complex Size in Living Cells Is Larger Than Expected
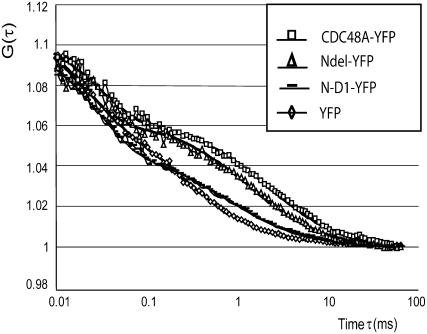
To investigate the size of CDC48A complexes in living cells, CDC48A-YFP constructs were expressed in Arabidopsis protoplasts. FCS was used to measure the diffusion times (τdif) of CDC48A-YFP proteins (full length, CDC48AA1A2, CDC48AN-D1, and CDC48ANdel) in the cytoplasm close to the PM and in the nucleus. To estimate cellular viscosities via the Stokes-Einstein equation (Eq. 5), the diffusion time of free YFP in cells was first measured. Differences in the diffusion times of protein molecules can point to mass differences between them and so the diffusion time of CDC48 in cells was measured. The results of the FCS measurements are based on nine independent transfections and summarized in Table II. In Figure 7, examples of autocorrelation curves are shown (both the raw data and fitted curves) for CDC48A-YFP, CDC48ANdel, CDC48AA1A2, and free YFP in the cytoplasm.
Table II.
Mean of the diffusion constant D (μm2/s) of various proteins, measured by FCS in the cytoplasm and nucleus of protoplasts
In the two-sample t test comparing CDC48A with one of the other proteins, the null hypothesis that the mean of the diffusion constants are the same is rejected when the absolute value of P is lower than the 5% significance level and accepted when P is higher. Values for D and rh were calculated via Equations 4 and 5. N-D1 is not expressed in the nucleus. sd is the sd of the D value; N is the number of curves.
| Protein | τdif | D | sd | N | t Test | rh |
|---|---|---|---|---|---|---|
| ms | μm2/s | nm | ||||
| In the cytoplasm | ||||||
| CDC48A | 1.886 | 3.5 | 0.8 | 20 | 20.4 | |
| YFP | 0.214 | 37.0 | 16.7 | 25 | P < 0.05 | 1.9 |
| N-D1 | 0.568 | 11.7 | 4.8 | 74 | P < 0.05 | 6.1 |
| Ndel | 1.323 | 6.0 | 1.7 | 18 | P < 0.05 | 11.9 |
| A1A2 | 1.223 | 6.2 | 0.7 | 8 | P < 0.05 | 11.4 |
| In the nucleus | ||||||
| CDC48A | 1.420 | 5.2 | 2.0 | 59 | 13.7 | |
| YFP | 0.222 | 30.4 | 8.0 | 30 | P < 0.05 | 2.3 |
| Ndel | 1.243 | 5.8 | 2.7 | 27 | P < 0.05 | 12.3 |
| A1A2 | 1.310 | 5.9 | 1.6 | 41 | P < 0.05 | 12.1 |
Figure 7.
Representative autocorrelation curves from CDC48A-YFP (squares), CDC48ANdel-YFP (triangles), and CDC48AN-D1-YFP (lines), compared to the curve for free YFP (lowest curve: diamonds) in Arabidopsis protoplasts. The autocorrelation curves were fitted with a one-component model. On the y axis the autocorrelation function G(τ) is depicted. The correlation time τ (ms) is depicted on the x axis in log scale.
First, the diffusion time of free YFP in protoplasts was determined in the cytoplasm close to the PM and in the nucleus. The average values for the diffusion times (τdif) for the cytoplasm and for the nucleus are listed in Table II. From the τdif values of each curve, the average diffusion coefficient D (μm2/s) and the hydrodynamic radius rh (nm) were calculated. The diffusion coefficients can be directly compared because, in contrast to the diffusion time, the diffusion coefficient is independent of the structural parameter or the shape of the volume. The hydrodynamic radius rh is a measure for the size of the protein (complex). The diffusion coefficient for free YFP in protoplasts was determined to be 37 μm2/s in the cytoplasm and 30 μm2/s in the nucleus. The calculated hydrodynamic radii were 1.9 nm and 2.3 nm, close to the theoretical rh for free YFP of 2.4 nm.
CDC48A-YFP in cells diffuses extremely slowly. A diffusion coefficient (D) of 3.5 μm2/s was found for CDC48A-YFP in the cytoplasm. To answer whether this is due to the hexameric nature of the CDC48A protein, the monomeric CDC48AN-D1-YFP mutant construct was also introduced into protoplasts. The diffusion coefficient of CDC48AN-D1-YFP was found to be 11.7 μm2/s, which is clearly faster than the CDC48A full-length protein. However, based on the crystal structure for VCP, CDC48A should have a rh of approximately 7 nm instead of the calculated 20.4 nm. Therefore, we hypothesized that the N domain, which is considered to be the domain responsible for most of the interactions of CDC48 (p97/VCP) with adaptor proteins like p47 (Dreveny et al., 2004), Ufd-1, and ubiquitinated proteins (Meyer et al., 2000), associated with some of these proteins in the full-length CDC48A as well as in the monomeric CDC48AN-D1-YFP. Therefore, we deleted the N domain from CDC48A and measured diffusion times of the mutant CDC48ANdel-YFP protein. Indeed, the diffusion coefficient of CDC48ANdel-YFP in the cytoplasm (6.0 μm2/s) is significantly higher than that of CDC48A (3.5 μm2/s). This could be explained by the lack of the interactive N domain. Unlike CDC48A, CDC48ANdel was not predominantly found in the hexameric form on native PAGE (Fig. 1B), which might also explain the faster diffusion of this mutant.
The diffusion coefficient of the CDC48AA1A2-YFP protein (6.2 μm2/s in the cytoplasm), the mutant which is reported to be severely delayed in hexamerization in vitro for p97/VCP (Wang et al., 2003a), was significantly higher than the diffusion coefficient of the CDC48A protein (3.5 μm2/s in the cytoplasm). Native PAGE showed that the CDC48AA1A2-YFP protein in cells is predominantly in the hexameric form, as well as CDC48A. Although the oligomerization of the CDC48A A1A2-YFP protein was not tested in the FRET-FLIM experiments, the FCS results suggest that fewer proteins associate with this mutant, resulting in faster diffusion of CDC48AA1A2-YFP. This implies that inhibition of ATP binding also inhibits complex formation with other proteins.
Surprisingly, no significant difference in diffusion coefficient between CDC48A-, CDC48AA1A2-, and CDC48ANdel-YFP was measured in the nucleus (Table II). In general, the diffusion coefficient in the nucleus was the same as in the cytoplasm for all proteins, except for CDC48A-YFP. The diffusion coefficient of CDC48A is much slower in the cytoplasm, suggesting that in contrast with the situation in the nucleus the protein is bound to a larger or different complex.
There is a discrepancy between the complex size estimated on native PAGE and the size calculated from the FCS analysis. An explanation might be that the CDC48A complex is associated with membranes or large molecules like the proteasome and therefore shows a very slow diffusion. For the native PAGE procedure, however, membranes are solubilized and connections of membranes with protein complexes can be lost.
From these data we propose that in vivo membrane-located CDC48A is found in larger protein complexes rather than in hexameric configuration only, while the N domain could be responsible for the interactions that lead to these large complexes.
DISCUSSION
The CDC48A protein was previously found to be one of the SERK1-interacting proteins (Rienties et al., 2005; Karlova et al., 2006). Because CDC48 proteins are reported to be active only in hexameric form, it was deemed essential to determine the oligomerization status in living cells. FRET-FLIM analysis indicated that the CDC48A-CrFP and CDC48A-YFP proteins are oligomerized in living cells. The CDC48A oligomers are predominantly hexameric, and the observed location of putative hexamers coincides with the interaction areas of SERK1 and CDC48A. These results suggest that the CDC48-CrFP/YFP proteins are indeed in their active conformation when expressed in living cells. Based on the prediction of this active configuration in vivo and the crystal structure of the related mammalian p97/VCP hexamer (DeLaBarre and Brunger, 2003), we used FRET-FLIM as a tool to confirm the presence of the hexameric configuration of CDC48A in vivo. Finally, the diffusion rates of the CDC48A proteins in vivo suggest that they are indeed part of a much larger oligomeric complex in vivo.
An unresolved and important question is what the functional relationship is between the CDC48A protein in an intact configuration and the SERK1 receptor. Active ATPases bind and hydrolyze ATP to release the energy for various processes in the cell, among which is the protein dislocation from the ER during ERAD (Jarosch et al., 2002). Many adaptor proteins were found to interact with the N domain of CDC48 or p97/VCP proteins, such as P47 (Dreveny et al., 2004), Shp1, and Ubx2 (Schuberth et al., 2004), Ufd1/Npl4 (Meyer et al., 2000), and Doa1 (Mullaly et al., 2006). These adaptor proteins recognize ubiquitinated substrates, thereby facilitating proteasome-dependent degradation. For many studies artificial substrates were used to study the ERAD system (Müller et al., 2005; DeLaBarre et al., 2006). However, to date hardly any physiological substrates of CDC48/VCP proteins are known, thereby limiting the understanding of the function of these proteins in general and in SERK1-mediated signaling in particular. Therefore, it could well be that the function of the interaction between CDC48A and SERK1 is to recognize and remove ubiquitinated receptors from the membrane, one of the proposals previously put forward by Aker et al. (2006). The IP3 receptor in ER membranes in Rat-1 fibroblasts was ubiquitinated upon ligand binding and coupled via the p97-Ufd1-Npl4 complex to proteasomal degradation (Alzayady et al., 2005).
DeLaBarre et al. (2006) found that two Args (Arg-586 and Arg-599) in the D2 domain of p97/VCP were essential for substrate interaction and ERAD, implicating that not every interaction with CDC48 proteins should go via the known adaptor proteins and the N terminus. This actually supports the idea that SERK1 interaction can occur with the C-terminal part of the CDC48A protein, as it has been found that way in yeast two-hybrid screens (Rienties et al., 2005). This was later confirmed by the FRET-FLIM evidence of interaction between SERK1-YFP and the C terminus of CDC48A (Aker et al., 2006). The result of this interaction was hypothesized to be trans-phosphorylation by the SERK1 kinase (Rienties et al., 2005). Only one phosphorylated residue of CDC48A was found (located in the N domain). Whether trans-phosphorylation of Ser-41 in the N domain of CDC48A is necessary for binding to the possibly ubiquitinated SERK1 receptor and subsequent degradation of the protein is still unclear. Binding of SERK1 to the C terminus could be part of the activation mechanism of the CDC48A protein complex. Rienties et al. (2005) showed that upon auto-phosphorylation of the SERK1-kinase domain, there was an increase in interaction with CDC48A. The mechanism of activation has still to be elucidated.
FRET-FLIM was also used to investigate the structure of CDC48A and one of its isomers, CDC48C, by calculating distances between parts of the molecule based on the FRET efficiencies. Differences in FRET efficiencies between CrFP and YFP, depending on whether they were fused to the N terminus of CDC48A or of CDC48C, led to the conclusion that the N domains of CDC48C are differently organized and probably closer together. It is not even clear whether CDC48C has a real N domain. It could be that CDC48C, being present only in the nucleus, is not involved in ubiquitin/proteasome-dependent proteolysis but has a function in nuclear envelope reformation during the exit from mitosis. This was also described for p97 (Hetzer et al., 2001). We therefore suggest that the “N domain” of CDC48C does not serve as a platform for binding ubiquitinated proteins and therefore probably does not protrude from the molecule.
Although FRET-FLIM is commonly used to estimate distances in multimeric complexes (Van Kuppeveld et al., 2002; Clayton et al., 2005), a cautionary remark should be made. Due to the hexameric structure and the lack of control over the precise composition of the contributing monomers in our model, the donor group CrFP and the acceptor group YFP are not at a fixed distance from each other. Each donor group could to a certain extent also interact with a different ensemble of acceptor groups at slightly different distances. It will require much more complex equations to deal with this situation, so the distances as calculated here may suggest a greater accuracy than actually can be measured. The distances calculated between the different subunits in the hexamer are nevertheless in close agreement with the distances as known for the structure of the homolog p97/VCP.
One of the surprising findings in our work was that the complex size of the CDC48A hexamer as derived from the diffusion coefficient determined by FCS was variable depending on the cellular compartment and also much larger than expected. The diffusion rate of CDC48A in the cytoplasm was significantly lower than in the nucleus, while the mobility of free YFP and the CDC48A mutants was comparable to that of these proteins in the nucleus. This suggested that the full-length hexameric CDC48A protein was the only functional protein and could therefore actively bind to other proteins and in the ER membrane compartments form complexes with adaptors and/or associate directly with the proteasomal complexes. Ubiquitination of the SERK1 receptor mediated by CDC48A might direct the receptor to the proteasome, but this has still to be elucidated. In the nucleus the CDC48A protein may have another function or is in a transit between targets and the proteasomal complexes.
Employing FRET-FLIM, we could provide details of SERK1 and CDC48A interaction and CDC48A oligomerization in vivo that would be difficult to resolve by other techniques. Using FCS it was shown that CDC48A hexamers in vivo are part of much larger complexes. Finally, we believe that the use of these microspectroscopic techniques is helpful to validate structural information of protein complexes in an intact cellular environment.
MATERIALS AND METHODS
Construction of the CrFP/YFP-Tagged Proteins
Plasmid pMON999 is a plant expression vector that contains the enhanced cauliflower mosaic virus 35S promoter, followed by a multiple cloning site and the terminator of the nopaline synthase gene (van Bokhoven et al., 1993). This plasmid, containing the cDNA of CrFP, enhanced YFP, or a fusion construct of CDC48A (At3g09840) or CDC48C (At3g01610), C-terminally linked to CrFP or YFP, was used for transfection of protoplasts.
Also, a mutant of CDC48A-CrFP and -YFP was constructed, the D2 deletion mutant (CDC48A-N-D1), missing the D2 domain from M448 to the end of the CDC48A sequence, by restriction with NcoI and subsequent ligation of the template.
A second mutant of CDC48A-YFP was constructed, missing the N domain (amino acids 1–188). Using the QuikChange XL site-directed mutagenesis kit, mutations K254T and K527T were introduced in the Walker A motifs of the D1 domain and the D2 domain, respectively. A combination of the two mutations, in a fusion protein with either CrFP or YFP, resulted in the A1A2 mutant. All constructs were checked by restriction enzyme analysis and the size of the protein was determined by western blotting. All constructs were verified by sequencing.
Protoplast isolation and transfection were performed as described before (Aker et al., 2006). FRET-FLIM was measured 16 h after transfections. For FCS measurements a low amount of fluorescent molecules in the detection volume is essential; therefore, cells were measured 6 h after transfections.
Protein Expression Analysis
Arabidopsis (Arabidopsis thaliana) mesophyll protoplasts were prepared and transfected according to a protocol described by Sheen (2001) with modifications, as described before (Aker et al., 2006). Sixteen to 20 h after transfection, protoplasts were lysed in RIPA buffer (50 mm Tris, pH 7.5, 150 mm NaCl, 1 mm MgCl2, 1% Nonidet P-40, containing a protease inhibitor tablet [Roche]) for 15 min on ice. Extracts were centrifuged for 7 min at 13,000 rpm at 4°C, and soluble fractions were supplemented with SDS-PAGE loading buffer, denatured at 95°C for 5 min before loading on an 8% gel, next to a prestained marker. Gels were blotted on nitrocellulose. For native PAGE, the soluble fractions were supplemented with native-PAGE loading buffer and loaded on a native gel of 4% to 8% acrylamide. Gels were blotted on PVDF membrane. The marker part of the blot was stained with Coomassie immediately after blotting. Anti-YFP was generated by Isogen, according to standard procedure. The serum was purified on a YFP-protA column. As a marker for native PAGE, a mixture of thyroglobuline (669 kD), ferritine (440 kD), and serum albumin (132 and 66 kD) was used.
In Vitro Trans-Phosphorylation and LC-MS/MS Analysis
For the in vitro trans-phosphorylation, 1 μg of SK1-KD was added to 1 μg of CDC48A in kinase buffer (Shah et al., 2001) plus 3 mm cold ATP and 1 μL of [γ32-P]ATP in a total reaction of 30 μL. After incubation of 30 min at 30°C, sample buffer was added. The sample was heated for 5 min at 95°C and loaded on SDS-PAGE. After drying the gel, the radioactivity was detected with a PhosphorImager using the ImageQuant program (Molecular Dynamics).
For MS analysis the in vitro phosphorylation was performed as described, without [γ32-P]ATP. The gel was not dried. The GST-CDC48A 125-kD bands were excised from the gel after staining with the Colloidal Coomassie staining kit (Invitrogen) and cut into 1-mm pieces. In-gel digestion was performed as described by Shevchenko et al. (1996). After destaining with water, Cys reduction (with 10 μL of 50 mm dithiothreitol in 50 mm NH4HCO3 for 1 h at 56°C), and alkylation (with 10 μL of 100 mm iodacetamide in 50 mm NH4HCO3 for 1 h at room temperature in the dark), the gel pieces were washed three times with 100 μL of 50 mm NH4HCO3 and dried in a vacuum centrifuge. For proteolytic digestion, the gel was treated overnight with 1 μL (in 10 μL of 50 mm NH4HCO3) trypsin (sequencing grade; Boehringer Mannheim). Finally, peptides were extracted from the gel pieces using 15 to 20 μL of 5% trifluoroacetic acid in water, followed by sonication for 1 min. A second gel extraction was done using 10 to 15 μL CH3CN/trifluoroacetic acid/water (25/1/84, v/v/v) mixture and sonication for 1 min. Extracted peptides were subjected to LC-MS/MS analysis. Each run with all spectra obtained was analyzed with Bioworks 3.2 (Thermo Electron). For identification of the peptides, the following proteins were added to a database; Arabidopsis CDC48A (P54609), the GST, trypsin (bovine, P00760), trypsin (porcin, P00761), keratin K2C1 (human, P04264), and keratin K1CI (human, P35527). The searches were done allowing Met oxidation and phosphorylation of Ser, Thr, and Tyr. The peptide identifications obtained were analyzed with Bioworks 3.2 with the following filter criteria: ΔCn > 0.08, Xcorr >2 for charge state 1+, Xcorr > 1.5 for charge state 2+, and Xcorr >3.3 for charge state 3+ as described previously (Peng et al., 2003).
FRET-FLIM
FRET takes place when two proteins coupled to appropriate fluorophores are in close distance from each other. FRET measurement is very sensitive since the rate of energy transfer is inversely proportional to the donor-acceptor distance to the power of 6 (Lakowicz, 1999). FRET can reveal distances between fluorophores that are typically in the range of 2 to 7 nm. FLIM is one method to measure FRET by estimating the reduction of CFP-donor lifetime in case of interaction with a YFP-acceptor molecule. FLIM was performed on a Bio-Rad Radiance 2100 MP system in combination with a Nikon TE 300 inverted microscope, as described by Russinova et al. (2004). From the fluorescence intensity image, the fluorescence lifetime is determined for each pixel. The full fluorescence decay per pixel was calculated for the donor molecule using a double exponential decay model. The fluorescence lifetime (τ value) can then be presented as a false-color image. First, the fluorescence lifetime of only the donor molecule was determined by analyzing all the measured pixels in the PM. The resulting average lifetime was subsequently used in the analysis where all decays were fit using two exponentials, with one lifetime fixed to the value for τ of the donor. This two-exponent model assumes two populations of donors: one that does not have an acceptor nearby and therefore has the unquenched lifetime τD, and one that has an acceptor near enough for energy transfer to occur and therefore has a reduced lifetime τDA. Fluorescence lifetime decays were fitted per pixel with a binning factor of 1. A double exponential decay model was used, with the lifetime of one component fixed to the value found for the unquenched lifetime of the donor. Fits were judged by the χr2 values (χr2 ≤ 1.1). The amount of photon counts in the peak channel was normally around 1,000. At least 15 cells were measured for each combination and location in two to five independent experiments. After measurements the distribution histograms of five representative cells were combined and plotted for each combination of proteins. Average lifetime per combination was determined and sds calculated. The distance between donor and acceptor were calculated via the following:
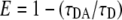 |
(1) |
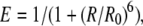 |
(2) |
where E is the transfer efficiency, R is the actual distance, and R0 is the Förster radius, indicating the distance where one observes 50% FRET efficiency. The R0 for the FRET pair CrFP and YFP was determined to be 4.9 nm, based on the excitation and emission spectra of the in vitro-produced proteins (J.W. Borst, unpublished data).
FCS
FCS makes use of a focused laser beam continuously illuminating a sample. Only the fluorescence from particles excited in a small observation volume is detected. The observation volume is defined by the focused laser beam in the x and y direction, and by the pinhole in the z direction, which makes monitoring of a subfemtoliter volume possible (Rigler et al., 1993). The entrance of a fluorophore into this volume gives rise to a burst of detected photons. The duration of this burst reflects the time the particle needs to diffuse through the observation volume, while the amplitude represents the molecular brightness. The intensity trace is correlated with itself at a later time point to yield the autocorrelation curve G(τ), from which the diffusion coefficient D and the average number of particles in the observation volume N can be obtained.
FCS measurements were performed on a Confocor 2-LSM 510 combination setup (Carl Zeiss). The 514-nm argon laser line was used to excite YFP (Hink et al., 2002). The system was calibrated with Rhodamine 6 Green solution (10 nm) and aligned using a rhodamine film. The FCS observation volume was positioned in the cytoplasm adjacent to the PM, to avoid measuring in chloroplasts. After a prebleach of 1 s with high (70% approximately 0.4 mW) laser power, three measurements of 20 s each were performed with low laser power (approximately 4 μW).
FCS data were analyzed using FCS Data Processor 1.4 (Shakun et al., 2005). Since measurements were performed in the cytoplasm, we fit the data with a model assuming free diffusion of protein and protein complexes in three dimensions, although interaction with the PM in some cases cannot be ruled out. An additional triplet-state term was used to describe the fast photophysics of YFP (e.g. conversion to a dark state, blinking).
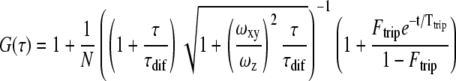 |
(3) |
Here, G(τ) is the autocorrelation function, N is the number of molecules in the detection volume, τdif is the diffusion time of the molecules, Ftrip is the fraction of molecules in the triplet state, and Ttrip is the triplet-state relaxation time. ωxy and ωz are the equatorial radius and axial radius of the Gaussian-shaped observation volume, respectively. The ratio ωz/ωxy is also called the structural parameter and was found to be between 5 and 10 in our measurements. The structural parameter was fixed to the value obtained with the Rhodamine 6 Green solution. The diffusion constants (D) were calculated from the τdif value for each curve and listed in a table for the PM and the nucleus. ses on the D value were calculated, and a two-sample t test with 95% significance was performed to compare the mean D value of CDC48A with the CDC48AA1A2, CDC48ANdel, and CDC48AN-D1 mutants.
The time a molecule on average spends in the observation volume τdif is linked to the translational diffusion coefficient D by Equation 4.
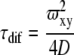 |
(4) |
The diffusion coefficient of a spherical particle is inversely proportional to the hydrodynamic radius rh according to the Stokes-Einstein relation (Edward, 1970):
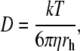 |
(5) |
in which k is the Boltzmann constant, T is the absolute temperature, and η is the viscosity of the solution. Assuming a globular particle, the molecular mass M is proportional to the hydrodynamic radius to the power of 3:
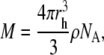 |
(6) |
where ρ is the mean density of the molecule and NA is Avogadro's number. Combination of Equations 4, 5, and 6 yields Equation 7, which shows that the diffusion constant scales only to the cubic root of the molecular mass and is independent of the observation volume.
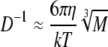 |
(7) |
Hence, the larger the molecule is, the lower the diffusion constant. To distinguish particles of different size with FCS, the diffusion times have to differ at least by a factor of 1.6, which corresponds to a 4-fold mass difference (Meseth et al., 1999).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers P54609 or At3g09840 (CDC48A) and Q9SS94 or At3g01610 (CDC48C).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Lifetime distributions of CDC48A.
Supplemental Figure S2. Lifetime distributions of CDC48C.
Supplemental Figure S3. Lifetime distributions of CDC48ANdel.
Supplementary Material
Acknowledgments
We thank Boudewijn van Veen for help in editing and formatting the images shown in this article, Isabella Nougalli and Adrie Westphal for help with the FCS measurements and calculations, and Sjef Boeren for help with the LC/MS measurements. We thank David Piston for the cerulean GFP variant.
This work was supported by the Dutch Organization for Research (grant no. ALW 812.06.004 to J.A.) and the Agrotechnology and Food Science Group of Wageningen University (J.W.B., Boudewijn van Veen, R.K., S.C.d.V.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Sacco C. de Vries (sacco.devries@wur.nl).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Aker J, Borst JW, Karlova R, de Vries S (2006) The Arabidopsis thaliana AAA protein CDC48A interacts in vivo with the somatic embryogenesis receptor-like kinase 1 receptor at the plasma membrane. J Struct Biol 156 62–71 [DOI] [PubMed] [Google Scholar]
- Alzayady KJ, Panning MM, Kelley GG, Wojcikiewicz RJ (2005) Involvement of the p97-Ufd1-NPl4 complex in the regulated endoplasmic reticulum-associated degradation of inositol 1,4,5-trisphosphate receptors. J Biol Chem 280 34530–34537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borst JW, Hink M, van Hoek A, Visser AJWG (2005) Effects of refractive index and viscosity on fluorescence and anisotropy decays of enhanced cyan and yellow fluorescent proteins. J Fluoresc 15 153–160 [DOI] [PubMed] [Google Scholar]
- Clayton AH, Walker F, Orchard SG, Henderson DF, Rothacker J, Nice EC, Burgess AW (2005) Ligand-induced dimer-tetramer transition during the activation of the cell surface epidermal growth factor receptor-A multidimensional microscopy analysis. J Biol Chem 280 30392–30399 [DOI] [PubMed] [Google Scholar]
- Davies JM, Tsuruta H, May AP, Weis WI (2005) Conformational changes of p97 during nucleotide hydrolysis determined by small-angle X-Ray scattering. Structure 13 183–195 [DOI] [PubMed] [Google Scholar]
- DeLaBarre B, Brunger AT (2003) Complete structure of p97/vasolin-containing protein reveals communication between nucleotide domains. Nat Struct Biol 10 856–863 [DOI] [PubMed] [Google Scholar]
- DeLaBarre B, Brunger AT (2005) Nucleotide dependent motion and mechanism of action of p97/VCP. J Mol Biol 347 437–452 [DOI] [PubMed] [Google Scholar]
- DeLaBarre B, Christianson JC, Kopito RR, Brunger AT (2006) Central pore residues mediate the p97/VCP activity required for ERAD. Mol Cell 22 451–462 [DOI] [PubMed] [Google Scholar]
- Dreveny I, Kondo H, Keij U, Shaw A, Zhang X, Freemont PS (2004) Structural basis of the interaction between the AAA ATPase p97/VCP and its adaptor protein p47. EMBO J 23 1030–1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edward JT (1970) Molecular volumes and Stokes-Einstein equation. J Chem Educ 47 261–265 [Google Scholar]
- Hetzer M, Meyer HH, Walther TC, Bilbao-Cortes D, Warren G, Mattaj JW (2001) Distinct AAA-ATPase p97 complexes function in discrete steps of nuclear assembly. Nat Cell Biol 3 1086–1091 [DOI] [PubMed] [Google Scholar]
- Hink MA, Bisseling T, Visser AJWG (2002) Imaging protein-protein interactions in living cells. Plant Mol Biol 50 871–883 [DOI] [PubMed] [Google Scholar]
- Huyton T (2003) The crystal structure of murine p97/VCP at 3.6 angstrom. J Struct Biol 144 337–348 [DOI] [PubMed] [Google Scholar]
- Indig FE, Partridge JJ, von Kobbe C, Aladjem MI, Latterich M, Bohr VA (2003) Werner syndrome protein directly binds to the AAA ATPase p97/VCP in an ATP-dependent fashion. J Struct Biol 146 251–259 [DOI] [PubMed] [Google Scholar]
- Jarosch E, Taxis C, Volkwein C, Bordallo J, Finley D, Wolf DH, Sommer T (2002) Protein dislocation from the ER requires polyubiquitination and the AAA-ATPase Cdc48. Nat Cell Biol 4 134–139 [DOI] [PubMed] [Google Scholar]
- Karlova R, Boeren S, Russinova E, Aker J, Vervoort J, de Vries S (2006) The Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASE1 protein complex includes BRASSINOSTEROID-INSENSITIVE1. Plant Cell 18 626–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakowicz JR (1999) Principles of Fluorescence Spectroscopy. Kluwer Academic/Plenum Publishers, New York
- Madeo F, Schlauer J, Zischka H, Mecke D, Frohlich KU (1998) Tyrosine phosphorylation regulates cell cycle-dependent nuclear localization of Cdc48p. Mol Biol Cell 1 131–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meseth U, Wohland T, Rigler R, Vogel H (1999) Resolution of fluorescence correlation measurements. Biophys J 76 1619–1631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer HH, Shorter JG, Seemann J, Pappin D, Warren G (2000) A complex of mammalian Ufd1 and NPl4 links the AAA-ATPase, p97, to ubiquitin and nuclear transport pathways. EMBO J 19 2181–2192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullaly JE, Chernova T, Wilkinson KD (2006) Doa1 is a Cdc48 adapter that possesses a novel ubiquitin binding domain. Mol Cell Biol 3 822–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller J, Piffanelli P, Devoto A, Miklis M, Elliot C, Ortmann B, Schulze-Lefert P, Panstruga R (2005) Conserved ERAD-like quality control of a plant polytopic membrane protein. Plant Cell 17 149–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Elias JE, Thoreen CC, Licklider LJ, Gygi SP (2003) Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LC-MS/MS) for large-scale protein analysis: the yeast proteome. J Proteome Res 2 43–50 [DOI] [PubMed] [Google Scholar]
- Pye VE, Dreveny I, Briggs LC, Sands C, Beuron F, Zhang X, Freemont PS (2006) Going through the motions: the ATPase cycle of p97. J Struct Biol 156 12–28 [DOI] [PubMed] [Google Scholar]
- Rancour DM, Dickey CE, Park S, Bednarek SY (2002) Characterization of AtCDC48A. Evidence for multiple membrane fusion mechanisms at the plane of cell division in plants. Plant Physiol 130 1241–1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rancour DM, Park S, Knight SD, Bednarek SY (2004) Plant UBX domain-containing protein 1, PUX1, regulates the oligomeric structure and activity of arabidopsis CDC48. J Biol Chem 279 54264–54274 [DOI] [PubMed] [Google Scholar]
- Rienties IM, Vink J, Borst JW, Russinova E, de Vries SC (2005) The Arabidopsis SERK1 protein interacts with the AAA ATPase AtCDC48A, the 14-3-3 GF14lambda and the PP2C phosphatase KAPP. Planta 221 394–405 [DOI] [PubMed] [Google Scholar]
- Rigler R, Mets U, Widengren J, Kask P (1993) Fluorescence correlation spectroscopy with high count rate and low background: analysis of translational diffusion. Eur Biophys J 22 169–175 [Google Scholar]
- Russinova E, Borst JW, Kwaaitaal M, Caňo-Delgado A, Yin Y, Chory J, deVries SC (2004) Heterodimerization and endocytosis of Arabidopsis brassinosteroid receptors BRI1 and AtSERK3 (BAK1). Plant Cell 16 3216–3229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah K, Vervoort J, de Vries SC (2001) Role of threonines in the Arabidopsis thaliana somatic embryogenesis receptor kinase 1 activation loop in phosphorylation. J Biol Chem 276 41263–41269 [DOI] [PubMed] [Google Scholar]
- Schuberth C, Richly H, Rumpf S, Buchberger A (2004) Shp1 and Ubx2 are adaptors of CDC48 involved in ubiquitin-dependent protein degradation. EMBO Rep 8 818–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheen J (2001) Signal transduction in maize and Arabidopsis mesophyll protoplasts. Plant Physiol 127 1466–1475 [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A, Wilm M, Vorm O, Mann M (1996) Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem 68 850–858 [DOI] [PubMed] [Google Scholar]
- Skakun VV, Hink MA, Digris AV, Engel R, Novikov EG, Apanasovich VV, Visser AJWG (2005) Global analysis of fluorescence fluctuation data. Eur Biophys J 34 323–334 [DOI] [PubMed] [Google Scholar]
- Tonaco IA, Borst JW, de Vries SC, Angenent GC, Immink RGH (2005) In vivo imaging of MADS-box transcription factor interactions. J Exp Bot 57 33–42 [DOI] [PubMed] [Google Scholar]
- van Bokhoven H, Verver J, Wellink J, van Kammen A (1993) Protoplasts transiently expressing the 200K coding sequence of cowpea mosaic virus B-RNA support replication of M-RNA. J Gen Virol 74 2233–2241 [DOI] [PubMed] [Google Scholar]
- Van Kuppeveld FJM, Melchers WJG, Willems PHGM, Gadella TWJ Jr (2002) Homomultimerization of the coxsackievirus 2B protein in living cells visualized by fluorescence resonance energy transfer microscopy. J Virol 76 9446–9456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Song C, Li CCH (2003. a) Hexamerization of p97-VCP is promoted by ATP binding to the D1 domain and required for ATPase and biological activities. Biochem Biophys Res Commun 300 253–260 [DOI] [PubMed] [Google Scholar]
- Wang Q, Song C, Yang X, Li CCH (2003. b) D1 ring is stable and nucleotide-independent, whereas D2 ring undergoes major conformational changes during the ATPase cycle of p97-VCP. J Biol Chem 35 32784–32793 [DOI] [PubMed] [Google Scholar]
- Woodman PG (2003) p97, a protein coping with multiple identities. J Cell Sci 116 4283–4290 [DOI] [PubMed] [Google Scholar]
- Zhang X, Shaw A, Bates PA, Newman RH, Gowen B, Orlova E, Gorman MA, Kondo H, Lally J, Leonard G, et al (2000) Structure of the AAA ATPase p97. Mol Cell 6 1473–1484 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.