Abstract
The distribution of resources between enzymes of photosynthetic carbon metabolism might be assumed to have been optimized by natural selection. However, natural selection for survival and fecundity does not necessarily select for maximal photosynthetic productivity. Further, the concentration of a key substrate, atmospheric CO2, has changed more over the past 100 years than the past 25 million years, with the likelihood that natural selection has had inadequate time to reoptimize resource partitioning for this change. Could photosynthetic rate be increased by altered partitioning of resources among the enzymes of carbon metabolism? This question is addressed using an “evolutionary” algorithm to progressively search for multiple alterations in partitioning that increase photosynthetic rate. To do this, we extended existing metabolic models of C3 photosynthesis by including the photorespiratory pathway (PCOP) and metabolism to starch and sucrose to develop a complete dynamic model of photosynthetic carbon metabolism. The model consists of linked differential equations, each representing the change of concentration of one metabolite. Initial concentrations of metabolites and maximal activities of enzymes were extracted from the literature. The dynamics of CO2 fixation and metabolite concentrations were realistically simulated by numerical integration, such that the model could mimic well-established physiological phenomena. For example, a realistic steady-state rate of CO2 uptake was attained and then reattained after perturbing O2 concentration. Using an evolutionary algorithm, partitioning of a fixed total amount of protein-nitrogen between enzymes was allowed to vary. The individual with the higher light-saturated photosynthetic rate was selected and used to seed the next generation. After 1,500 generations, photosynthesis was increased substantially. This suggests that the “typical” partitioning in C3 leaves might be suboptimal for maximizing the light-saturated rate of photosynthesis. An overinvestment in PCOP enzymes and underinvestment in Rubisco, sedoheptulose-1,7-bisphosphatase, and fructose-1,6-bisphosphate aldolase were indicated. Increase in sink capacity, such as increase in ADP-glucose pyrophosphorylase, was also indicated to lead to increased CO2 uptake rate. These results suggest that manipulation of partitioning could greatly increase carbon gain without any increase in the total protein-nitrogen investment in the apparatus for photosynthetic carbon metabolism.
The steady-state biochemical model of leaf photosynthesis developed by Farquhar et al. (1980), with subsequent developments (von Caemmerer, 2000; Farquhar et al., 2001), is immensely valuable in linking biochemical properties of photosynthesis, in particular, those of Rubisco, with in vivo photosynthetic rates. The steady-state photosynthesis model (Farquhar et al., 1980) provides an elegant way to represent photosynthesis under stable environmental conditions. While its simplicity and robustness are huge strengths, because the Farquhar et al. (1980) model is based on limitation by either Rubisco or regeneration of ribulose-1,5-bisphosphate (RuBP), it does not provide a framework for scaling from the emergent areas of photosynthetic genomics, proteomics, and metabolomics. Further, photosynthesis in nature is rarely at steady state. For example, there is great temporal and spatial heterogeneity of light and temperature within plant canopies (Pearcy, 1990; Zhu et al., 2004a). The dynamics of photosynthesis following environmental perturbations has been studied for many years (Pearcy, 1983, 1990; Walker, 1992; Pearcy et al., 1997). For example, in the 1980s transient processes of photosynthesis, in particular, “induction” of O2 evolution from isolated chloroplasts and oscillations observed at saturating [CO2], became the object of modeling because understanding oscillations means understanding photosynthesis (Walker, 1992). Complex dynamic models of photosynthesis with different levels of detail have been built (Laisk and Walker, 1986, 1989; Pettersson and Ryde-Pettersson, 1988; Laisk et al., 1989; Giersch et al., 1990; Woodrow and Mott, 1993; Pearcy et al., 1997; Pettersson, 1997; Fridlyand et al., 1999; Fridlyand and Scheibe, 1999a, 1999b; Giersch, 2000; Poolman et al., 2000). These models contributed to the understanding of control of different transient properties of photosynthesis, such as the role of extrachloroplastic phosphate concentration (Pettersson and Ryde-Pettersson, 1988), the importance of the ATP/NADPH stoichiometry in initiating and sustaining oscillations (Laisk et al., 1991, 1992b), the necessity for the nonphotochemical quenching of excitation at PSII (Laisk et al., 1992a), and the importance of turnover time (or poolsizes) of the Calvin cycle intermediates in regulating metabolism (Fridlyand and Scheibe, 1999a). Recently, Laisk and Edwards (2000) constructed a model of C4 photosynthesis, which proposed a new understanding of the accumulation of CO2 in the bundle sheath cells, demonstrating the heuristic power of complex models.
As with steady-state models of photosynthesis, dynamic models hold great application potential. In addition to being used as tools for hypothesis testing regarding the mechanisms of different in vivo dynamic behavior, photosynthesis models have also been used to calculate the flux control coefficient (CC) of enzymes for CO2 uptake rate (A; e.g. Poolman et al., 2000). A dynamic model of the Calvin cycle has also been used to identify targets to engineer to increase photosynthesis (Poolman, 1999). Modern biotechnology now has the power to engineer each enzyme in photosynthesis and to alter resource allocation between the enzymes. The challenge now is to determine which changes or combination of changes may be most beneficial. Application of control analysis to dynamic models of the Calvin cycle (e.g. Pettersson and Ryde-Pettersson, 1988; Poolman et al., 2000) has identified several enzymes with high flux CC, suggesting that increasing the amounts of these enzymes would increase the maximum photosynthetic rate. However, there may be little scope to add more protein to the chloroplast stroma or photosynthetic cell (Pyke and Leech, 1987; Zhu et al., 2004b), and little desire in contemporary agriculture to add yet more nitrogen to crops. Given a fixed resource of total protein-nitrogen available to the enzymes of photosynthetic carbon metabolism, increases in the amounts of some enzymes will inevitably require compensatory decrease in concentrations of other enzymes. There are a total of 38 enzymes involved in photosyntehic carbon metabolism, including the Calvin cycle, photorespiratory metabolism (PCOP), starch synthesis, and Suc synthesis. Even if a single step increase or decrease in each enzyme was considered, identification of the optimal protein-nitrogen distribution into different enzymes would involve testing >109 permutations. In reality, a continuous variation in the amount of each enzyme should be considered, generating even more permutations. Clearly, experimental transgenic manipulation could not cope with so many permutations. Modern computational power and optimization tools now offer a means toward such a guide. In this particular study, we used an evolutionary algorithm (Goldberg, 1989) as a tool to study the optimization of partitioning of protein-nitrogen between the enzymes of photosynthetic carbon metabolism.
The aim of this study was to demonstrate the application of dynamic photosynthesis models in engineering higher light-saturated photosynthetic rates. Another aim was to determine whether total protein, expressed as protein-nitrogen, is partitioned optimally with respect to maximizing light-saturated photosynthetic rate for a typical C3 leaf and, if not, what reallocation would maximize light-saturated photosynthesis. Under current atmospheric [CO2] and [O2], photorespiration can decrease photosynthesis by up to 30% (Long and Drake, 1991; Zhu et al., 2004b). Therefore, inclusion of the photorespiratory pathway is critical to any model of photosynthetic carbon metabolism addressing photosynthetic efficiency in normal air. Apparently, no current dynamic model of photosynthetic carbon metabolism includes all the reactions in the Calvin cycle, photorespiratory metabolism, starch synthesis, and Suc synthesis. We have therefore extended earlier models of photosynthetic carbon metabolism (Pettersson and Ryde-Pettersson, 1988; Laisk et al., 1989; Poolman et al., 2000) with new knowledge about the enzymes, and have added all enzyme-catalyzed reactions of the photorespiratory or PCOP pathway, and starch and Suc anabolism (Fig. 1). The specific objectives of this study were 2-fold. (1) To produce a complete model of photosynthetic carbon metabolism (including PCOP) that could mimic dynamic phenomena closely associated with the presence of photorespiration. These include testing its ability to quantitatively achieve photosynthetic rates and steady-state metabolite levels similar to those observed in vivo and qualitatively mimic observed responses to perturbations, specifically, a transient decrease in oxygen, both under inorganic phosphate-limiting and -nonlimiting conditions. (2) To test the hypothesis that photosynthetic rate may be increased by altering the partitioning of resources between the different enzymes of photosynthetic carbon metabolism, without increasing the total amount of protein-nitrogen.
Figure 1.
Reactions of the Calvin cycle, starch synthesis, photosynthetic carbon oxygenation pathway, triose-P export, and Suc synthesis represented in the model (Supplemental Appendix S1, A and B). The numbers in the figure identify the reactions as they are represented in each equation. The number in subscript for each reaction rate (vm) is the reaction number shown in this diagram. Double-headed arrows indicate reversible reactions and single-headed arrows indicate essentially irreversible reactions. Reactions above the double dotted line are those of the chloroplast stroma and those below of the cytosol. The circles in the chloroplast membrane represent translocators: for triose-P-inorganic phosphate and glycolate-glycerate exchange. The metabolites in different compartments, i.e. cytosol and stroma, are represented separately in the model. For example, FBP in the stroma is treated as separate from FBP in the cytosol.
RESULTS
A was simulated as the solution of the complete set of linked differential equations representing concentration change of each metabolite in photosynthetic carbon metabolism (Fig. 1; all abbreviations are listed in Table I). A first test of the model was its ability to simulate light-saturated A versus ci response. The model was allowed to run to a steady-state A for each ci. A brief damped oscillation was observed when the simulation was initiated (e.g. Fig. 2A). From these simulations a typical A/ci response was constructed (Fig. 2B) that showed a biphasic increase in A, as predicted from the Farquhar et al. (1980) model. With a lower [O2], a higher initial slope of A versus ci was simulated (Fig. 2B). The predicted A is within the range of photosynthetic rates typically observed in vivo (e.g. Long et al., 1996). Given a high sink capacity (i.e. high activity of cytosolic Fru-1,6-bisphosphatase [FBPase]), decrease in [O2] led to an increase in A as expected due to inhibition of photorespiration (Fig. 2C). When the maximum rate of cytosolic FBPase was decreased by 90% to mimic sink limitation, a progressive decrease in A was simulated. Here, lowering [O2] to 2% decreased rather than increased A (Fig. 2D) as it does when sink capacity is not limiting (Fig. 2C). This counterintuitive decrease in A upon decrease in [O2] (Fig. 2D) corresponds to in vivo observations of phosphate-limited photosynthesis (Sharkey, 1985). Phosphate-limited photosynthesis is due to decreased availability of phosphate for ATP synthesis, which in turn slows synthesis of RuBP and correspondingly CO2 carboxylation.
Table I.
Compiled abbreviations
Abbreviations defined in the text or used in the figures or the supplemental files are listed below.
| Abbreviationa | Full Name | Unitsb |
|---|---|---|
| A | Leaf CO2 uptake rate | μmol m−2 s−1 |
| ADPG | ADP-Glc | mmol L−1 |
| ADPGPP | ADP-Glc pyrophosphorylase | NA |
| ATc | Sum of ADP and ATP concentrations in the cytosol | mmol L−1 |
| [CO2] | CO2 concentration in the atmosphere or stroma | μmol mol−1 or mmol L−1 |
| CA | Sum of ADP and ATP concentrations in the stroma | mmol L−1 |
| CN | Sum of NADP+ and NADPH concentrations in the stroma | mmol L−1 |
| CP | Sum of organic and inorganic phosphate in the stroma | mmol L−1 |
| CPc | Sum of organic and inorganic phosphate in the cytosol | mmol L−1 |
| DHAP | Dihydroxyacetone-P | mmol L−1 |
| DPGA | 1,3-Bisphosphoglycerate | mmol L−1 |
| E4P | Erythrose-4-P | mmol L−1 |
| Et | Total Rubisco concentration | mmol L−1 |
| F6P | Fru-6-P | mmol L−1 |
| FBP | Fru-1,6-bisphosphate | mmol L−1 |
| F26BP | Fru-2,6-bisphosphate | mmol L−1 |
| G1P | Glc-1-P | mmol L−1 |
| G6P | Glc-6-P | mmol L−1 |
| GAP | Glyceraldehyde-3-P | mmol L−1 |
| GAPDH | Glyceraldehyde-3-P dehydogenase | NA |
| GCA | Glycolate | mmol L−1 |
| GCEA | Glycerate | mmol L−1 |
| GDC | Gly decarboxylase | NA |
| GLUc | Glutamate | mmol L−1 |
| GLYc | Gly in cytosol | mmol L−1 |
| GOA | Glyoxylate | mmol L−1 |
| HexP | Hexose-P, includes F6P, G6P, and G1P | NA |
| GGAT | Gly glyoxylate aminotransferase | NA |
| GSAT | Glyoxylate Ser aminotransferase | NA |
| HPR | Hydroxypyruvate | mmol L−1 |
| KGc | α-Ketoglutarate | mmol L−1 |
| ODE | Ordinary differential equation | NA |
| [O2] | Oxygen concentration in atmosphere | mmol mol−1 |
| OPOP | Pyrophosphate | mmol L−1 |
| PCOP | Photosynthetic carbon oxidation pathway | NA |
| PCRC | Photosynthetic carbon reduction cycle | NA |
| PenP | Sum of concentrations of Ri5P, Ru5P, and Xu5P | mmol L−1 |
| 3-PGA | 3-Phosphoglycerate | mmol L−1 |
| PGCA | 3-Phosphoglycolate | mmol L−1 |
| PRK | Ribulose-5-P kinase | NA |
| PGCA Pase | Phosphoglycolate phosphatase | NA |
| PGA kinase | 3-Phosphoglycerate kinase | NA |
| Ri5P | Rib-5-P | mmol L−1 |
| PRK | Phosphoribulose kinase | mmol L−1 |
| Ru5P | Ribulose-5-P | mmol L−1 |
| Rubisco | Ribulose-1,5-bisphosphate carboxylase/oxygenase | NA |
| Rt | Total RuBP concentration in the stroma | mmol L−1 |
| RuBP | Ribulose-1,5-bisphosphate | mmol L−1 |
| S7P | Sedoheptulose-7-P | mmol L−1 |
| SBP | Sedoheptulose-1,7-bisphosphate | mmol L−1 |
| SBPase | Sedoheptulosebisphosphatase | NA |
| SERc | Ser in the cytosol | mmol L−1 |
| SPP | Suc-P phosphatase | NA |
| SPS | Suc-P synthetase | NA |
| TPU | Triose-P utilization | NA |
| SUCc | Suc in the cytosol | mmol L−1 |
| SUCPc | Suc-P in the cytosol | mmol L−1 |
| T3P | Triose-P, including DHAP and GAP | mmol L−1 |
| v111 | The rate of RuBP oxygenation | μmol m−2 s−1 or mmol L−1 s−1 |
| UDPGc | Uridine diphosphate Glc | mmol L−1 |
| UDPGP | UDP-Glc pyrophosphorylase | NA |
| UT | Total UDP and UTP concentration in the cytosol | mmol L−1 |
| v1 | The rate of RuBP carboxylation | μmol m−2 s−1 or mmol L−1 s−1 |
| vatpg | The rate of ATP formation in the cytosol | μmol m−2 s−1 or mmol L−1 s−1 |
| vpga_use | The rate of PGA utilization in the cytosol | μmol m−2 s−1 or mmol L−1 s−1 |
| Xu5P | Xylulose-5-P | mmol L−1 |
A suffix c was added to the name of a metabolite appearing in the cytosol. For example, PGA is phosphoglycerate in the stroma, while PGAc is phosphoglycerate in the cytosol.
NA, Not applicable.
Figure 2.
Responses of net leaf A to different environmental perturbations, as predicted by the model. The response of A to ci was examined in silico by mimicking the experiment as it might be conducted in vivo. In the simulation, ci was initially set at 700 μmol mol−1. When steady-state A was achieved, ci was decreased by 70 μmol mol−1, A recorded at steady state, and then ci decreased again until 70 μmol mol−1 was reached. The photosynthetic photon flux density, represented by the maximum rate of ATP synthesis in this model, was assumed to be 15 mmol L−1 s−1, which corresponds to 450 μmol m−2 s−1 on a leaf-area basis. A shows oscillations when simulation was initiated at a [CO2] of 700 μmol mol−1. B shows the A/ci response based on the steady-state A achieved at each ci used in the simulation. The solid circles and solid triangles represent A/ci at 2% and 21% [O2], respectively. C, A further numerical experiment simulated in silico the effect of changing [O2] on A. The model was initiated at a ci of 280 μmol mol−1, and after 2,000 s the [O2] was decreased from 21% to 2%. The system was restored to 21% after a further 2,000 s. D repeats the numerical experiment of C, but here the activity of cytosolic FBPase was reduced by 90% to simulate sink limitation. In all simulations temperature is set to 25°C and light assumed to be saturating. The initial concentrations of the metabolites used in the model were based on reported levels for light-adapted leaves (table C3 in Supplemental Appendix S1C, table D3 in Supplemental Appendix S1D, and table E3 in Supplemental Appendix S1E).
Figure 3 is the same experiment as Figure 2, C and D, but shows the underlying dynamics in metabolite concentrations. These simulations were initiated with concentrations of metabolites in “typical” C3 leaves during light-saturated photosynthesis obtained via a literature analysis (see tables C3, D3, and E3 in Supplemental Appendix S1). On initiation, rapid transients in concentrations were predicted that stabilized within about 60 s to a steady state (Fig. 3). The steady-state concentrations were of the same magnitude as the initial concentrations (Fig. 3; compare with tables C3, D3, and E3 in Supplemental Appendix S1). Once steady state was obtained, the system was perturbed by lowering [O2] to 2% to inhibit RuBP oxygenation. Despite this marked perturbation of the balance between Calvin cycle and PCOP, the system quickly restabilized to a new steady state, again within 60 s (Fig. 3). On returning to 21% [O2], concentrations returned to the original steady state (Fig. 3), demonstrating the stability of the system of ordinary differential equations (ODEs) and the method of numerical integration. Sensitivity analysis of every kinetic parameter for every enzyme showed that parameters for Rubisco and sedoheptulose-1,7-bisphosphatase (SBPase) were by far the most critical in determining the simulated A (Table II; Supplemental Appendix S1, C–E).
Figure 3.
Change in the concentration of four metabolites in the simulations reported in Figure 2, C and D. A and E, [RuBP]; B and F, [T3P]; C and G, [T3Pc]; and D and H, [PGA]; A to D are for the simulation (Fig. 2C) assuming a high capacity for cytosolic FBPase and E to H for the simulation (Fig. 2D) assuming a low capacity for cytosolic FBPase, simulating sink limitation of photosynthesis. See Table I for definitions of the abbreviations used in this figure.
Table II.
The concentrations of the enzymes of carbon metabolism for the initial and optimized leaves
These are the absolute concentrations of the enzymes represented in relative terms in Figure 5A. The CCs for the initial and optimized leaves as predicted by the evolutionary algorithm are listed.
| Enzyme Name | Initial Concentrations of Enzymes Used in the Simulation | CC for the Initial Nitrogen Distribution | Concentrations of Enzymes for the Optimized Leaf | CC for the Optimized Nitrogen Distribution |
|---|---|---|---|---|
| mg N m−2 | mg N m−2 | |||
| Rubisco | 517 | 0.72 | 795 | 0.88 |
| PGA kinase | 12.2 | 0.00002 | 5.06 | 0.006 |
| GAP dehydrogenase | 68.8 | 0.0002 | 75.0 | 0.72 |
| FBP aldolase | 6.42 | 0.005 | 11.7 | 0.015 |
| FBPase | 25.5 | 0.004 | 35.9 | 0.044 |
| Transketolase | 34.9 | 0.008 | 18.4 | 0.022 |
| SBP aldolase | 6.21 | 0.003 | 7.43 | 0.0078 |
| SBPase | 1.29 | 0.8 | 4.90 | 0.006 |
| Phosphoribulose kinase | 7.64 | 0.006 | 8.55 | 0.01 |
| ADPGPP | 0.49 | 0.0001 | 4.88 | 0.0058 |
| Phosphoglycolate phosphatase | 85.2 | <0.000001 | 1.42 | 0.0014 |
| Glycerate kinase | 6.36 | <−0.000001 | 1.31 | <0.000001 |
| Glycolate oxidase | 4.77 | <0.000001 | 1.49 | −0.03 |
| Ser glyoxylate aminotransferase | 17.3 | <0.000001 | 3.03 | −0.017 |
| Glycerate dehydrogenase | 2.64 | <−0.000001 | 0.778 | <0.000001 |
| Glu glyoxylate aminotransferase | 21.8 | <−0.000001 | 4.47 | −0.01 |
| GDC | 179 | <−0.000001 | 18.6 | −0.14 |
| Cytosolic FBP aldolase | 0.566 | 0.005 | 0.280 | 0.00072 |
| Cytosolic FBPase | 2.24 | 0.004 | 1.44 | 0.0017 |
| UDP-Glc pyrophosphorylase | 0.0748 | 0.004 | 0.0718 | 0.00087 |
| Suc-P synthetase | 0.202 | 0.014 | 0.153 | <0.000001 |
| Suc-P phosphatase | 0.129 | −0.0002 | 0.07.3 | <0.000001 |
| FBPase | 0.0207 | 0.001 | 0.0101 | <0.000001 |
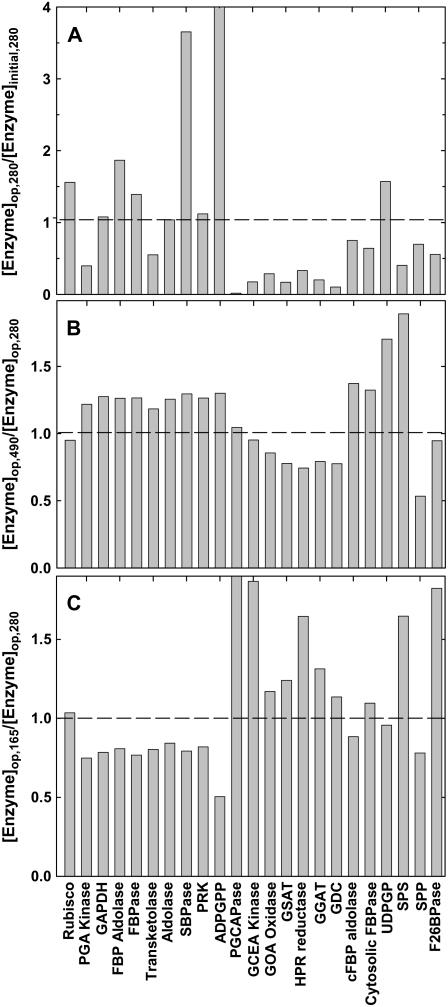
Optimization of nitrogen allocation between the enzymes of photosynthetic carbon metabolism using an evolutionary algorithm suggested that light-saturated photosynthesis could be substantially increased (Fig. 4). Over generations of numerical selection, a progressive increase in investment in some enzymes at the expense of others was observed (Fig. 4). During the simulation, the total protein-nitrogen concentration available for the enzymes of the photosynthetic carbon metabolism was kept constant. After 1,500 generations, when the maximum A was reached, simulated A had increased 76% over initial A. Rubisco, SBPase, ADP-Glc pyrophosphorylase (ADPGPP), and Fru-1,6-bisphosphate (FBP) aldolase were increased, whereas enzymes in the PCOP pathway and reactions involved in Suc synthesis were decreased (Table II; Fig. 5A). In the next application of the evolutionary algorithm, we used the optimized nitrogen allocation selected at an intercellular [CO2] (ci) of 280 μmol mol−1, reflecting the current atmospheric concentration of 380 μmol mol−1. We then reran the algorithm for a ci of 490 μmol mol−1, which corresponds to the ambient [CO2] of 700 μmol mol−1 that is anticipated for the end of this century. At this future CO2, this simulated “evolution” moved nitrogen away from Rubisco and the enzymes in the photorespiratory metabolism, whereas all other enzymes in the Calvin cycle, ADPGPP, and cytosolic enzymes involved in Suc synthesis were increased (Fig. 5B). Simulated “evolution” at a low [CO2], i.e. a ci of 165 μmol mol−1 corresponding to the assumed average ci of the past 25 million years, reallocated nitrogen to Rubisco and enzymes of photorespiratory metabolism from other enzymes in the Calvin cycle and ADPGPP (Figs. 5C and 6C). When capacities for 3-phosphoglycerate (PGA), glyceraldehyde-3-P (GAP), and dihydroxyacetone-P (DHAP) export via the phosphate translocator were increased, to simulate increased sink capacity, the optimal concentrations of enzymes under both current (280 μmol mol−1), elevated (490 μmol mol−1), and past [CO2] (165 μmol mol−1) were largely unaltered compared to those obtained with low export rates of PGA, GAP, and DHAP (Fig. 5 versus Fig. 6). An exception is UDP-Glc pyrophosphorylase, which was increased when export capacity was high (Fig. 6). This implies that to achieve high A when capacity for triose-P export is high, increase in capacity for both starch synthesis, as suggested by the increase in ADPGPP, and Suc synthesis, as suggested by the increase in UDP-Glc phosphorylase, is necessary.
Figure 4.
Selection of the optimal partitioning of resources between the enzymes of carbon metabolism to maximize light-saturated photosynthetic rate (A). A, Increase in A over 1,500 generations of selection using an evolutionary algorithm. B to F show the changes in the amounts of five of these enzymes relative to their initial values over these 1,500 generations. Selection was at light saturation and a ci of 280 μmol mol−1. Total protein in the enzymes of carbon metabolism was fixed at 1 g m−2 of leaf area.
Figure 5.
The change in concentrations of the enzymes of carbon metabolism selected over 1,500 generations. A, The ratio of the enzyme concentrations optimized by the evolutionary algorithm at a ci of 280 μmol mol−1 ([Enzyme]op,280) compared to the initial concentrations ([Enzyme]initial,280); B, the ratio of the optimal enzyme concentration in a ci of 490 μmol mol−1 ([Enzyme]op,490) compared to the optimum in a ci of 280 μmol mol−1. C, The ratio of the optimal enzyme concentration in a ci of 165 μmol mol−1 ([Enzyme]op,165) compared to the optimum in a ci of 280 μmol mol−1. The maximal rate of triose-P (PGA, GAP, and DHAP) export is 1 mmol L−1 s−1. Selection in each generation is for the maximum light-saturated A, as in Figure 4. Definitions of the abbreviations for these enzymes are given in Table I.
Figure 6.
As for Figure 5, but with a triose-P (PGA, GAP, and DHAP) maximum export rate of 3 mmol L−1 s−1.
Following the Farquhar et al. (1980) model, we estimated the maximal Rubisco carboxylation rate (Vcmax) and maximal electron transfer rate (Jmax) for the initial and optimized leaf based on the Michaelis-Menten constants for CO2 and O2 used in this study. The Vcmax and Jmax for the initial leaf, i.e. generation 0, were 75 and 116 μmol m−2 s−1, respectively. The Vcmax and Jmax were 121 and 192 μmol m−2 s−1, respectively, for the optimized leaf under the current ci of 280 μmol mol−1.
DISCUSSION
The objectives of this study were (1) to produce a complete model of photosynthetic carbon metabolism that could mimic dynamic phenomena associated with the presence of photorespiration, and (2) to test the hypothesis that photosynthetic rate may be increased by altering the partitioning of resources between the different enzymes of photosynthetic carbon metabolism without increasing the total amount of protein-nitrogen.
A dynamic model of photosynthetic carbon metabolism that includes all reactions involved in the Calvin cycle, photorespiratory pathway (PCOP), starch synthesis, and Suc synthesis was developed by extending previous models (Pettersson and Ryde-Pettersson, 1988; Laisk et al., 1989; Poolman et al., 2000). The model was shown to achieve a stable and realistic rate of light-saturated A, and qualitatively reproduced well-established dynamics of A following environmental perturbations, specifically, alteration of intercellular [CO2] and [O2], where cytosolic phosphate was either in surplus or was limiting to triose-P transfer from the chloroplast to cytosol. The model as used here assumed maximum activation of the enzymes of photosynthetic carbon metabolism, commensurate with light saturation, so avoiding many feedback controls imposed by thioredoxin, pH, Mg2+, and Rubisco activation via its activase. Despite this simplification, a steady-state A was attained, and a new steady state was quickly reestablished following perturbation of [O2]. Furthermore, the original steady-state A was reattained when the system was returned to 21% [O2] from 2% [O2] (Fig. 2, C and D). The well-established response of steady-state A to intercellular [CO2] (A/ci) was also successfully predicted (Fig. 2B). Equally, steady-state concentrations of key intermediates in silico were similar to reported in vivo concentrations (Fig. 3 versus tables C3, D3, and E3 in Supplemental Appendix S1). In theory, a series of so many (38) linked differential equations would have many potential steady-state solutions due to the nonlinearity of the rate equations with respect to the substrate and product involved in the reaction (Supplemental Appendix S1A). In our simulations, only one was attained, and this same solution was regained after a transient perturbation of the system by changing [O2]. The simulated values of A, 16 μmol m−2 s−1 in normal air and 18 μmol m−2 s−1 in 2% [O2], are within the range typical for healthy C3 leaves (Wullschleger, 1993). Given the assumed model structure (Fig. 1), the steady-state concentrations are purely a function of the kinetic parameters of the enzymes and the conserved quantities (see “Materials and Methods,” section “Equations for Conserved Quantities”). When the model was initiated with metabolite concentrations extracted from the literature, an initial damped oscillation was observed in the metabolite concentrations, but the steady-state values were all within an order of magnitude of the initial literature values (Fig. 3). This implies that the many controls and feedbacks within the Calvin cycle and associated carbon metabolism are not essential to obtaining a stable high rate of photosynthesis, but may of course affect the steady-state A and may affect the stability of the system when light is not saturating. Sensitivity analysis showed that simulated A was most sensitive to variation in the kinetic parameters of Rubisco and SBPase, while hardly effected (<0.5%) by a 5% variation in most other enzymes (Table II; Supplemental Appendix S1, C–E). This may explain why the model produces surprisingly realistic values and responses of A despite the disparate sources of data for parameterization. Kinetic parameters for Rubisco and SBPase are among the most studied and best defined. Further, use of antisense technology to alter enzyme amounts, coupled with flux analysis, has similarly shown Rubisco and SBPase to dominate control of light-saturated photosynthesis (Raines, 2003).
The calculated CCs for different enzymes of carbon metabolism with the “evolved” optimal nitrogen allocation were not identical (Table II). Previous theoretical analysis had shown that under a constant total protein-nitrogen available, the optimal flux CCs for different enzymes in a pathway are not identical but depend on the pathway structure and enzyme properties, such as molecular masses, catalytic numbers, Michaelis-Menten constants, etc. (Heinrich and Klipp, 1996). Intuitively, if one enzyme has a large molecular mass and a small catalytic number, a relatively large increase in nitrogen investment will be required to gain a relatively small increase in the activity of this enzyme. Given a fixed total pool of leaf protein-nitrogen, the increase of nitrogen to this enzyme will inevitably decrease nitrogen available to all other enzymes, which leads to a decrease in flux. Another possibility for the unequal CCs for different enzymes is that the evolutionary algorithm finds a local rather than global optimum, in the same way that real evolution can move into a situation from which it cannot easily escape. This would occur when reversion is through nonviable phenotypes. An example in real evolution might be the oxygenase activity of Rubisco. To check this possibility, we repeated the “evolution” three times, and the results obtained were almost identical even though random selection was reseeded on each run.
While a decrease in [O2] normally increases A by lowering the amount of carbon entering PCOP relative to the Calvin cycle, experimentally it has been shown that this does not apply when export of carbohydrate from the chloroplast is limited by cytosolic phosphate, which inhibits ATP synthesis (Sharkey, 1985). Again, the system faithfully simulated this peculiar phenomenon of photosynthesis. When a low activity of cytosolic FBPase, corresponding to a low sink strength, was simulated, A at 2% [O2] was lower than at 21% [O2], converse to the observation when a high activity of cytosolic FBPase, corresponding to a high sink strength, was simulated (Fig. 2, C and D). This qualitatively mimics the response observed in vivo (Sharkey, 1985). In addition, our model successfully simulated the previously reported counterintuitive effects of SBPase on A under different sink strengths (Poolman et al., 2001), i.e. increase in SBPase activity led to increased A under high sink strength but decreased A under low sink strength (data not shown). These tests showed that the system of linked differential equations and the method of numerical integration produced quantitatively realistic solutions and qualitatively realistic behavior. This was critical to progress to the second objective of the study, i.e. to determine whether protein-nitrogen is allocated optimally with respect to light-saturated photosynthetic rate and, if not, what reallocation would maximize light-saturated photosynthesis.
Redistribution of nitrogen among different enzymes could in theory lead to a large increase in A, more than 76% from 16 to 28 μmol m−2 s−1 (Fig. 4). Relative to the initial concentration of enzymes, increases in Rubisco, FBP aldolase, SBPase, and ADPGPP were required to increase the maximum A (Fig. 5A). The increases in these enzymes are consistent with the high flux CCs that these enzymes have (Stitt et al., 1991; Poolman et al., 2000). Intriguingly, SBPase is one enzyme where an approximately 10% increase in A and productivity has been observed in transgenics overexpressing SBPase (Lefebvre et al., 2005; Tamoi et al., 2006). This simulation suggests that further gains could be achieved if, in addition, Rubisco, FBP aldolase, and ADPGPP (or together with UDP-Glc phosphorylase when the membrane triose-P export capacity is high) were overexpressed.
These increases were achieved at the expense of enzymes of photorespiratory metabolism and of storage carbohydrate metabolism downstream of triose-P. The fact that the actual distribution does not match the modeled optimal distribution could have many explanations. Assuming that the kinetic properties of the enzymes used approximate to reality and that these enzymes are largely activated at light saturation, then an apparently nonoptimal partitioning could have four potential causes.
(1) Evolution in the natural environment selects for those individuals producing the maximum number of viable progeny, while the evolutionary algorithm selected for maximal light-saturated A at 25°C. Survival is by definition critical to maximizing progeny produced, and selection for survival may sometimes be counter to photosynthetic efficiency. For example, the simulation selected for very large decreases in all photorespiratory (PCOP) enzymes in order to maximize A. Consistent with this prediction, 60% decreases in the amounts of both Gly decarboxylase (GDC) and Ser glyoxylate aminotransferase have been found not to affect A (Wingler et al., 1997, 1999). However, under high-temperature stress, e.g. 40°C, the flux of carbon into PCOP would increase 2.5 times relative to 25°C, based on the kinetic properties of Rubisco (calculated from Farquhar et al. [1980], using the temperature functions of Bernacchi et al. [2001]). If the leaf is simultaneously water stressed such that the intercellular [CO2] is decreased, then this flux will be even larger. Under these conditions, enzyme limitation of the rate at which carbon entering the PCOP is returned back to the Calvin cycle could be lethal, so this apparent overinvestment in enzymes of PCOP may be to ensure survival under stress at the expense of a higher A under optimal conditions. This suggests that for crops grown in environments where few stresses are likely, e.g. cool temperate moist climates and some protected environments, engineering the changes suggested by Figure 5A could result in large increases in leaf photosynthesis. Such increases in leaf photosynthesis can be closely linked to increase in crop productivity (for review, see Long et al., 2006). The cost, though, may be decreased stress tolerance.
(2) Another reason why the concentration of enzymes in the PCOP has not decreased as predicted here may be because the ratio of RuBP oxygenation to carboxylation is fixed at given CO2 and O2 concentrations. If enzyme concentrations in the PCOP decrease and Rubisco specificity remains constant, metabolites in the PCOP will inevitably accumulate (Zhu et al., 2004b). Unless plants evolve mechanisms to effectively utilize the accumulated metabolites, this carbon will be lost from the Calvin cycle and may slow regeneration of RuBP. One potential solution is to increase the Rubisco specificity to CO2 versus O2, correspondingly lowering enzymes in the photorespiratory pathway.
(3) An increase in the activity of SBPase is predicted. Why has normal evolution not already selected this increase? SBPase affects the branch between regeneration of RuBP and starch synthesis (Woodrow and Berry, 1988). Its activity is known to be modulated by many other metabolites, critically, thioredoxins, pH, and Mg2+ (Raines et al., 1999, 2000; Schurmann and Jacquot, 2000). SBPase could only act as a key point of regulation if its activity is sufficiently low to exert control on flux through the Calvin cycle. Its lower than predicted amount (Fig. 5A) might be argued to reflect an important control function. However, the prediction of the evolutionary algorithm is supported by two independent studies in which SBPase activity has been transgenically increased by expression of the gene from either Arabidopsis (Arabidopsis thaliana) or Chlamydomonas in tobacco (Nicotiana tabacum). Both manipulations gave large increases in A and final dry matter yield, without any apparent detrimental phenotype (Lefebvre et al., 2005; Tamoi et al., 2006). Increase in SBPase activity by 4.3-fold increased light-saturated photosynthesis by 23% and final dry matter production by 50% (Tamoi et al., 2006). Our simulation assumes a constant NADPH and so assumes no limitation in electron transport. Increase in SBPase therefore allows a large increase in the rate of regeneration of RuBP, reflected in a 65% increase in Jmax. Recent experimental analyses suggest that control of Jmax may be colimited by the capacity of the cytochrome bf complex and SBPase (Price et al., 1998; Harrison et al., 2001). So, while increase in SBPase may result in an increase in A at light saturation, this may be moderated by limitation in whole-chain electron transport.
(4) Atmospheric [CO2] has risen from approximately 270 μmol mol−1 in 1850 to 384 μmol mol−1 today. Yet, our current C3 plants evolved over the past 25 million years in a [CO2] of 235 μmol mol−1 (Barnola et al., 2003). Because CO2 competitively inhibits RuBP oxygenation at Rubisco, the flux of carbon into photorespiration as a proportion of the flux into photosynthesis gradually decreases with increase in [CO2]. Calculations using this model showed that the proportions of flux into photorespiration are 30%, 26.1%, and 18.3% for atmospheric [CO2] of 235, 270, and 380 μmol mol−1, respectively. Therefore, the flux into the photorespiratory pathway was decreased about one-third with increase of atmospheric [CO2] from 270 to 380 μmol mol−1 since the industrial revolution. Thus, they may be poorly adapted to the increase that has occurred in the brief period of 150 years on an evolutionary time scale. Consistent with this, the optimized enzyme distributions under a higher ci, i.e. 490 μmol mol−1, showed the lower concentration of photorespiratory enzymes needed to deal with the decreased flux of carbon into this pathway with rising [CO2] (Fig. 5B).
In conclusion, a complete model of photosynthetic carbon metabolism that is capable of simulating photosynthesis in normal air, i.e. in the presence of photorespiration, was developed. It demonstrates the potential of combining dynamic models of metabolic pathways with evolutionary algorithms to identify the combinations of changes most likely to lead to increased productivity. Application is currently limited by the lack of complete published analyses of all photosynthetic carbon metabolism proteins for a single leaf. As a result, the exact percentage of increase in each enzyme might be taken as a trend rather than an absolute numerical value. Despite these limitations, application of an evolutionary algorithm suggests that partitioning of resources between proteins is suboptimal with respect to maximizing productivity. While some of the inferred improvements, in particular, increase in SBPase, have been identified experimentally, other suggested improvements are subtle. The most important inference is that very substantial gains in photosynthetic productivity could be obtained by altered partitioning of resources, with importance for future crop production in the absence of severe stress and in adapting crop photosynthesis to global atmospheric change.
MATERIALS AND METHODS
Four Stages of Model Development
The model was developed in four stages. (1) Rate equations for each discrete step in photosynthetic carbon metabolism were developed based on literature and standard equations for enzyme kinetics. (2) Equations for conserved quantities were developed where the sum of two or more metabolites is a conserved quantity, e.g. [NADPH] + [NADP+] is a constant. (3) Differential equations were developed to describe the rate of concentration change in each metabolite. Each differential equation describes the consumption and production of a metabolite. These equations were linked to form the complete model. (4) Algorithms for solving the system of linked differential equations were developed. Because there is no analytical solution for the system of differential equations describing photosynthetic carbon metabolism, computationally efficient algorithms for numerical integration that could deal with very different rates of concentration change in different metabolite pools were identified to solve the system of differential equations.
(1) Rate Equations
The complete set of rate equations is presented in Supplemental Appendix S1A. The reactions in C3 photosynthetic carbon metabolism are numbered in the diagrammatic representation of the linked pathways where a number is given for each enzyme-catalyzed reaction (Fig. 1; all abbreviations are listed in Table I). The numbers in this figure were used as subscripts in the notation of different reaction rates, e.g. v2, v3, etc. In the diagram, reversible reactions are indicated by double-headed arrows and essentially irreversible reactions are indicated as a single-headed arrow pointing from the substrate to the product. The reactions were categorized into equilibrium and nonequilibrium reactions. The reactions assumed to be in equilibrium (Laisk et al., 1989) were interconversion between the following: (1) GAP and DHAP in both stroma and cytosol; (2) xylulose-5-P (Xu5P), Rib-5-P (Ri5P), and ribulose-5-P (Ru5P); and (3) Fru-6-P (F6P), Glc-6-P (G6P), and Glc-1-P (G1P; Fig. 1).
The following equations were used to describe DHAP ↔ GAP in the stroma:
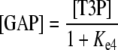 |
(1.1.1) |
 |
(1.1.2) |
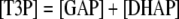 |
(1.1.3) |
where Ke4 is the equilibrium constant of the reaction and [T3P] represents the sum of [GAP] and [DHAP].
The relationship between the concentrations of (1) Ri5P, Ru5P, Xu5P, and their sum (PenP), and (2) F6P, G6P, G1P, and their sum (HexP) were derived similarly (Supplemental Appendix S1A).
All nonequilibrium reactions, except four, were assumed to obey Michaelis-Menten kinetics, modified as necessary for the presence of inhibitor(s) or activator(s). For a general reversible reaction of the form A + B ↔ C + D, the rate equation used was:
 |
(1.1.4) |
following the standard kinetic equation for a reversible reaction with two substrates and two products (Cleland, 1963), where [A], [B], [C], and [D] represent the concentrations of the metabolites A, B, C, and D, respectively. KmA, KmB, KmC, and KmD are the Michaelis-Menten constants for A, B, C, and D, respectively. ke is the equilibrium constant of this reaction; Vm is the maximum rate of this reaction.
For a general nonreversible reaction, A + B → C + D, the generalized rate equation was:
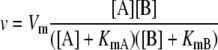 |
(1.1.5) |
after Segel (1975).
The presence of a competitive inhibitor (E) changes the apparent Michaelis-Menten constant of the corresponding substrate (Segel, 1975). For example, in the presence of a competitive inhibitor of metabolite A in a nonreversible reaction A + B → C + D, the rate equation used was modified for the model here to:
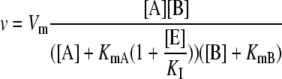 |
(1.1.6) |
where KI is the inhibition constant.
These generic equations were used to describe, as appropriate, the enzyme-catalyzed steps of the Calvin cycle, starch synthesis, triose-P export, Suc synthesis, and the PCOP (Supplemental Appendix S1A). The only exceptions, where reaction order or conditions required specific formulations, were the reactions catalyzed by Rubisco, ADPGPP, the chloroplast envelope phosphate translocator, and GDC (Supplemental Appendix S1A).
The concentration of Rubisco active sites in the chloroplast stroma is of the same order of magnitude as the concentration of the substrate RuBP (Bassham and Krause, 1969; Dietz and Heber, 1984; Schimkat et al., 1990; Woodrow and Mott, 1993). Taking account of the high Rubisco concentration, Farquhar (1979) developed an equation relating the rates of carboxylation and oxygenation to total RuBP concentration (Rt). The solution of this equation is closely approximated by:
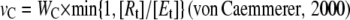 |
(1.1.7) |
where WC is calculated as:
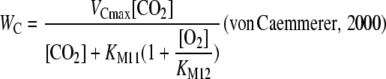 |
(1.1.8) |
where VCmax represents the maximum rate of RuBP carboxylation; KM11 the Michaelis-Menten constant of CO2; and KM12 the Michaelis-Menten constant of O2. Some intermediates of the Calvin cycle other than RuBP bind to the Rubisco active site and competitively inhibit RuBP carboxylation (Badger and Lorimer, 1981). This is implemented in the model as a modification of Equation 1.1.8 (compare with Farquhar, 1979; von Caemmerer, 2000):
 |
(1.1.9) |
where Kr is the Michaelis-Menten constant for RuBP, where KI11, KI12, KI13, KI14, and KI15 are the respective constants for PGA, FBP, sedoheptulose-1,7-bisphosphate (SBP), phosphate, and NADPH inhibition of RuBP binding to Rubisco active sites (Badger and Lorimer, 1981). To maintain the activity of Rubisco, Rubisco activase is required (Portis, 1995, 2003). The ratio of Rubisco to Rubisco activase was assumed to be unity (Eckardt et al., 1997). Effectively, Rubisco and Rubisco activase always accompany each other and have the same molar concentrations in this model.
The reactions catalyzed by ADPGPP (Eq. 1.1.10) and the phosphate translocator (Eqs. 1.1.11–1.1.14) were assumed to conform to the rate equations developed by Pettersson and Ryde-Pettersson (1988).
 |
(1.1.10) |
 |
(1.1.11) |
 |
(1.1.12) |
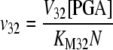 |
(1.1.13) |
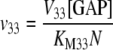 |
(1.1.14) |
We assumed that the rate equations for all reactions in PCOP followed Michaelis-Menten kinetics. GDC is an enzyme complex that includes four different component proteins (P protein, H protein, T protein, and L protein), which function together to catalyze the oxidative decarboxylation and deamination of Gly resulting in the formation of CO2, NH3, and the concomitant reduction of NAD+ to NADH (Douce et al., 2001). These four proteins act in concert. As a simplification, one Michaelis-Menten equation representing the first reaction in GDC, the Gly decarboxylation reaction, was used to represent the overall reactions catalyzed by GDC. Ser is a competitive inhibitor of Gly in the decarboxylation reaction (Oliver and Raman, 1995); therefore, the rate equation of Gly decarboxylation was:
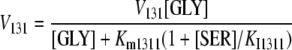 |
(1.1.15) |
where V131 represents the maximum rate of Gly decarboxylation; [GLY] and [SER] represent the concentrations of Gly and Ser in the cytosol; Km1311 represents the Michaelis-Menten constant for Gly; and KI1311 represents the constant for competitive inhibition of GDC by Ser.
Our model made further assumptions about PCOP metabolism. First, there was no limitation to metabolite transport from mitochondrion to peroxisome, i.e. spatial differentiation between peroxisome and mitochondrion were not considered. Second, the transfer of glycerate and glycolate between stroma and cytosol was assumed to follow Michaelis-Menten kinetics (Howitz and McCarty, 1985a, 1985b, 1986) with the rate equations incorporating glycerate competitive inhibition of the binding of glycolate and vice versa (Howitz and McCarty, 1985b, 1986).
The rate equations used for describing all reactions in this model of photosynthetic C3 carbon metabolism are listed in Supplemental Appendix S1A. This model needs a large number of constants and parameters, such as Michaelis-Menten constants, inhibition constants, and the maximal enzyme activities. No consistent set of constants/parameters for any single plant species was available. As a compromise, these constants/parameters were obtained by surveying peer-reviewed studies for different species. Given this compromise, whenever possible, we used constants/parameters from spinach (Spinacia oleracea) in the model. All Michaelis-Menten constants and inhibition constants for the Calvin cycle, starch synthesis, the PCOP reactions, and Suc synthesis are listed in table C1 of Supplemental Appendix S1C, table D1 of Supplemental Appendix S1D, and table E1 of Supplemental Appendix S1E, respectively.
Similarly, there is no consistent set of maximum enzyme activities for all the enzymes in photosynthetic carbon metabolism yet. As a compromise, we developed a standardized set of maximum enzyme activities for a “typical” C3 leaf under high light based on the literature survey. To do this, we first compiled the activities of enzymes involved in the Calvin cycle, photorespiratory pathway, and Suc synthesis pathways as reported in the literature. The enzyme activities in each pathway were then normalized relative to the Vm of Rubisco as this was the enzyme most commonly measured. If Rubisco was not available, then the ratio to either glycolate oxidase or Suc-P synthetase was used, and then corrected to Rubisco based on the average ratio of these two enzymes to Rubisco from other studies. During the normalization, the ratio of Vm of each enzyme in the pathway to that of the chosen enzyme was calculated. The normalization step was done for Vm values from each study and then averaged across all studies (tables C2, D2, and E2 of Supplemental Appendix S1, C–E). These ratios were used as the basis for building the Vm for each enzyme used in the model.
Finally, to construct the “typical” C3 leaf used in this study, we assumed that the total protein-nitrogen in enzymes of the photosynthetic carbon metabolism is 1 g m−2. The mass of nitrogen in each enzyme in 1 m2 leaf area was then calculated based on the number of active sites, catalytic rate per active site, molecular mass of each enzyme, and the ratios between Vm of different enzymes. Mole of each protein is then calculated based on the molecular mass and the mass of each protein. To convert activity to a volume basis, the volume of stroma and cytosol was calculated assuming a leaf chlorophyll concentration of 1 g chlorophyll m−2 and 30 μL stroma mg−1 chlorophyll and 30 μL cytosol mg−1 chlorophyll (Harris and Koniger, 1997). The Vm for each enzyme was then calculated based on the amount of each enzyme and the volume of the compartment that it occupies in 1 m2 leaf area.
(2) Equations for Conserved Quantities
During photosynthesis, the total concentration of the adenylate nucleotides ([CA]) in the chloroplast stroma, i.e. the sum of [ATP] and [ADP], was assumed to remain constant. It was assumed that AMP does not participate in photosynthetic carbon metabolism and that its concentration is negligible, following Pettersson and Ryde-Pettersson (1988) and Poolman et al. (2000). Similarly, the sum of [NADPH] and [NADP] in the chloroplast stroma ([CN]) was assumed a constant. The export of PGA, GAP, or DHAP from the chloroplast to the cytosol is associated with a counterimport of phosphate, mediated by a phosphate translocator (Fig. 1). Therefore, the total concentration of phosphate in the stroma ([CP]) was also assumed constant (Fliege et al., 1978). Three equations represent these conserved sums:
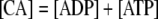 |
(1.2.1) |
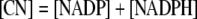 |
(1.2.2) |
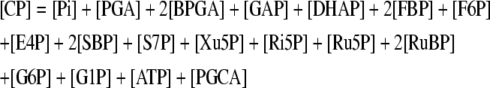 |
(1.2.3) |
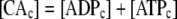 |
(1.2.4) |
 |
(1.2.5) |
 |
(1.2.6) |
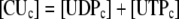 |
(1.2.7) |
(3) The Differential Equations
The rate of change in concentration of a metabolite is represented by the difference between the rate(s) of the reaction(s) generating the metabolite and the rate(s) of the reaction(s) consuming the metabolite. For example, RuBP is generated from the phosphorylation of Ru5P, catalyzed by Ru5P kinase, with a reaction rate v13, and RuBP is consumed by oxygenation (v111) and carboxylation (v1; Fig. 1). Thus, the rate of RuBP concentration change is:
 |
(1.3.1) |
The volume of the chloroplast stroma can be different from that of the cytosol in a typical higher plant cell (Winter et al., 1993, 1994; Leidreiter et al., 1995), with the ratio between the volume of the stroma and that of the cytosol varying in different species and under different conditions (Winter et al., 1993, 1994; Leidreiter et al., 1995). As a simplification, a ratio of 1:1 was assumed in calculating concentrations in the two compartments (Supplemental Appendix S1B). The differential equations describing rates of change of all metabolites are listed in Supplemental Appendix S1B.
(4) Algorithms for Solving the System Representing the C3 Photosynthetic Carbon Metabolism
The complete set of linked differential equations (Supplemental Appendix S1B) forms a system of ODEs. This system of ODEs, together with the rate equations (Supplemental Appendix S1A), forms a complete description of photosynthetic carbon metabolism. After initial testing of alternative numerical integration routines, we chose the ode15s procedure of MATLAB (version 6; MathWorks) to solve the system of ODEs (Supplemental Appendix S1B). The solution of the ODEs provided by ode15s is the time evolution of the concentrations of metabolites. This routine was chosen because of its computational efficiency in dealing with a “stiff” set of equations, i.e. where rates of change differ significantly between components. The initial concentrations of metabolites were parameterized based on “typical” literature values for concentrations in illuminated leaves (table C3 in Supplemental Appendix S1C, table D3 in Supplemental Appendix S1D, and table E3 in Supplemental Appendix S1E). All simulations assumed that photosynthesis was light saturated, with all light-regulated enzymes fully activated and a constant NADPH level of 1 mm, i.e. this assumes that any consumption of NADPH is instantaneously replenished.
Experiments
(1) Model Validation
To test the utility and stability of the model, the following numerical experiments were conducted. The model was run: (1) to determine if stable and realistic steady-state levels of metabolites and A can be obtained; and (2) to determine if by perturbing the system, particularly, with respect to the balance between PCOP and Calvin cycle, by lowering [O2] from 21% to 2%, a new stable steady-state could be obtained and that the former steady-state solution was regained on return to 21% [O2]. This test was conducted with and without inorganic phosphate limitation, i.e. with high and low chloroplast triose-P export capacities.
(2) Development and Application of an Evolutionary Algorithm to Identify Optimal Distribution of Enzymes Given Fixed Protein-Nitrogen Investments
It was assumed that the amount of enzyme is proportional to Vm. An evolutionary algorithm (Goldberg, 1989) was developed to select for the maximal light-saturated A given a fixed total nitrogen investment in all the enzymes of photosynthetic carbon metabolism. A similar approach was used to study the optimal morphology of plants on land (Niklas, 1999). An evolutionary algorithm uses mechanisms of biological evolution: reproduction, mutation, natural selection, and survival of the fittest. Candidate sets of the enzyme concentrations, i.e. solutions to the optimization problem, play the role of individuals in a population and select for higher rates of light-saturated A. In this study, the starting distribution of protein-nitrogen among the enzymes was obtained as described above. For the second and subsequent generations, the concentrations of all enzymes were mutated through random variation according to:
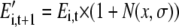 |
(1.5.1) |
where Ei,t+1′ is the unadjusted concentration of ith enzyme in the metabolism network at the (t + 1)th generation, Ei,t is the concentration of ith enzyme in the metabolism network at tth generation, and N(x, σ) represents a normally distributed random number of mean x and sd σ. In this particular study, we assumed that x is 0 and σ is 0.05. When the new concentration of each enzyme had been computed, the total was summed and then the amount of each enzyme was corrected by its product with the ratio of the original total and new to ensure that the total amount of protein remains constant.
In each generation, production of 16 new individuals was simulated. Each individual, representing a set of enzymes required for the model of photosynthetic carbon metabolism, was used as input for the model to simulate the steady-state light-saturated photosynthesis. The individual with the highest A was selected and used to “seed” the next generation. This was repeated for 1,500 generations when a steady optimized solution is identified. After the optimal nitrogen allocation was identified for a ci of 280 μmol mol−1, the optimized nitrogen allocation was used as the initial enzyme concentration to reoptimize for a ci of 490 or 165 μmol mol−1, which corresponds to the assumed ci for atmospheric [CO2] in the year 2100 (Prentice et al., 2001) and the average [CO2] of the past 25 million years (Barnola et al., 2003). The ci to ca ratio was assumed to be constant at 0.7 (Wong et al., 1979). The simulations were conducted twice, first with a high maximum chloroplast membrane triose-P export capacity (3 mmol L−1 s−1) and second with a low maximum capacity (1 mmol L−1 s−1).
The flux CC for the maximal velocity of each enzyme used in the system was calculated. For example, to calculate CC for Rubisco maximum carboxylation velocity (V1), we decreased V1 by 5% and then examined the percentage change in A (Δ%A). The CC of V1 was then estimated as:
 |
(1.5.2) |
This same procedure was repeated to calculate the response coefficient (Kacser et al., 1995) for all kinetic parameters, such as Michaelis-Menten constants, for each enzyme. These CCs are listed in Table II and response coefficients are listed in Supplemental Appendix S1, C to E.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Appendix S1. The appendices include the equations and parameter values used in the article.
Supplementary Material
This work was co-supported by the National Center for Supercomputing Applications at the University of Illinois and the U.S. National Science Foundation (grant no. IBN 04–17126).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Stephen P. Long (stevel@life.uiuc.edu).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Badger MR, Lorimer G (1981) Interaction of sugar phosphate with the catalytic site of RuBP-carboxylase. Biochemistry 20 2219–2225 [DOI] [PubMed] [Google Scholar]
- Barnola JM, Raynaud D, Lorius C, Barkov NI (2003) Historical CO2 record from the Vostok ice core. In Trends: A Compendium of Data on Global Change. Carbon Dioxide Information Analysis Center, Oak Ridge National Laboratory, Department of Energy, Oak Ridge, TN
- Bassham JA, Krause GH (1969) Free energy changes and metabolic regulation in steady state photosynthetic carbon reduction. Biochim Biophys Acta 189 207–221 [DOI] [PubMed] [Google Scholar]
- Bernacchi CJ, Singsaas EL, Pimentel C, Portis JAR, Long SP (2001) Improved temperature response functions for models of Rubisco-limited photosynthesis. Plant Cell Environ 24 253–259 [Google Scholar]
- Cleland WW (1963) The kinetics of enzyme-catalyzed reactions with two or more substrates or products. Biochim Biophys Acta 67 104–137 [DOI] [PubMed] [Google Scholar]
- Dietz KJ, Heber U (1984) Rate-limiting factors in leaf photosynthesis. 1. Carbon fluxes in the Calvin cycle. Biochim Biophys Acta 767 432–443 [Google Scholar]
- Douce R, Bourguignon J, Neuburger M, Rébeillé F (2001) The glycine decarboxylase system: a fascinating complex. Trends Plant Sci 6 167–176 [DOI] [PubMed] [Google Scholar]
- Eckardt NA, Snyder GW, Portis AR, Ogren WL (1997) Growth and photosynthesis under high and low irradiance of Arabidopsis thaliana antisense mutants with reduced ribulose-1,5-bisphosphate carboxylase/oxygenase activase content. Plant Physiol 113 575–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farquhar GD (1979) Models describing the kinetics of ribulose biphosphate carboxylase-oxygenase. Arch Biochem Biophys 193 456–468 [DOI] [PubMed] [Google Scholar]
- Farquhar GD, von Caemmerer S, Berry JA (1980) A biochemical model of photosynthetic CO2 assimilation in leaves of C3 species. Planta 149 78–90 [DOI] [PubMed] [Google Scholar]
- Farquhar GD, von Caemmerer S, Berry JA (2001) Models of photosynthesis. Plant Physiol 125 42–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fliege R, Flugge U-I, Werner K, Heldt HW (1978) Specific transport of inorganic phosphate, 3-phosphoglycerate and triose phosphates across the inner membrane of the envelope in spinach chloroplasts. Biochim Biophys Acta 502 232–247 [DOI] [PubMed] [Google Scholar]
- Fridlyand L, Backhausen J, Scheibe R (1999) Homeostatic regulation upon changes of enzyme activities in the Calvin cycle as an example for general mechanisms of flux control. What can we expect from transgenic plants? Photosynth Res 61 227–239 [Google Scholar]
- Fridlyand LE, Scheibe R (1999. a) Controlled distribution of electrons between acceptors in chloroplasts: a theoretical consideration. Biochim Biophys Acta 1413 31–42 [Google Scholar]
- Fridlyand LE, Scheibe R (1999. b) Regulation of the Calvin cycle for CO2 fixation as an example for general control mechanism in metabolic cycles. Biosystems 51 79–93 [DOI] [PubMed] [Google Scholar]
- Giersch C (2000) Mathematical modelling of metabolism. Curr Opin Plant Biol 3 249–253 [PubMed] [Google Scholar]
- Giersch C, Lammel D, Farquhar GD (1990) Control analysis of photosynthetic CO2 fixation. Photosynth Res 42 75–86 [DOI] [PubMed] [Google Scholar]
- Goldberg DE (1989) Genetic Algorithms in Search, Optimization and Machine Learning. Addison-Wesley, Reading, MA
- Harris GC, Koniger M (1997) The ‘high’ concentration of enzymes within the chloroplast. Photosynth Res 54 5–23 [Google Scholar]
- Harrison EP, Olcer H, Lloyd JC, Long SP, Raines CA (2001) Small decreases in SBPase cause a linear decline in the apparent RuBP regeneration rate, but do not affect Rubisco carboxylation capacity. J Exp Bot 52 1779–1784 [DOI] [PubMed] [Google Scholar]
- Heinrich R, Klipp E (1996) Control analysis of unbranched enzymatic chains in states of maximal activity. J Theor Biol 182 243–252 [DOI] [PubMed] [Google Scholar]
- Howitz KT, McCarty RE (1985. a) Kinetic characteristics of the chloroplast envelope glycolate transporter. Biochemistry 24 2645–2652 [Google Scholar]
- Howitz KT, McCarty RE (1985. b) Substrate specificity of the pea chloroplast glycolate transporter. Biochemistry 24 3645–3650 [Google Scholar]
- Howitz KT, McCarty RE (1986) d-Glycerate transport by the pea chloroplast glycolate carrier. Studies on [1-C]d-glycerate uptake and d-glycerate dependent O2 evolution. Plant Physiol 80: 390–395 [DOI] [PMC free article] [PubMed]
- Kacser H, Burns JA, Fell DA (1995) The control of flux. Biochem Soc Trans 23 341–366 [DOI] [PubMed] [Google Scholar]
- Laisk A, Edwards GE (2000) A mathematical model of C-4 photosynthesis: the mechanism of concentrating CO2 in NADP-malic enzyme type species. Photosynth Res 66 199–224 [DOI] [PubMed] [Google Scholar]
- Laisk A, Eichelmann H, Oja V, Eatherall A, Walker DA (1989) A mathematical model of the carbon metabolism in photosynthesis. Difficulties in explaining oscillations by fructose 2,6-bisphosphate regulation. Proc R Soc Lond B Biol Sci 237 389–415 [Google Scholar]
- Laisk A, Oja V, Heber U (1992. a) Steady-state and induction kinetics of the photosynthetic electron-transport related to donor side oxidation and acceptor side reduction of photosystem-1 in sunflower leaves. Photosynthetica 27 449–463 [Google Scholar]
- Laisk A, Oja V, Walker D, Heber U (1992. b) Oscillations in photosynthesis and reduction of photosystem-1 acceptor side in sunflower leaves—functional cytochrome b6/f-photosystem 1 ferredoxin-NADP reductase supercomplexes. Photosynthetica 27 465–479 [Google Scholar]
- Laisk A, Siebke K, Gerst U, Eichelmann H, Oja V, Heber U (1991) Oscillations in photosynthesis are initiated and supported by imbalances in the supply of ATP and NADPH to the Calvin cycle. Planta 185 554–562 [DOI] [PubMed] [Google Scholar]
- Laisk A, Walker DA (1986) Control of phosphate turnover as a rate-limiting factor and possible cause of oscillations in photosynthesis: a mathematical model. Proc R Soc Lond B Biol Sci 227 281–302 [Google Scholar]
- Laisk A, Walker DA (1989) A mathematical model of electron transport. Thermodynamics necessity for photosystem II regulation: ‘Light stroma’. Proc R Soc Lond B Biol Sci 237 417–444 [Google Scholar]
- Lefebvre S, Lawson T, Zakhleniuk OV, Lloyd JC, Raines CA (2005) Increased sedoheptulose-1,7-bisphosphatase activity in transgenic tobacco plants stimulates photosynthesis and growth from an early stage in development. Plant Physiol 138 451–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leidreiter K, Kruse A, Heineke D, Robinson DG, Heldt HW (1995) Subcellular volumes and metabolite concentrations in potato (Solanum tuberosum cv Desiree) leaves. Bot Acta 108 439–444 [Google Scholar]
- Long SP, Drake BG (1991) Effect of the long-term elevation of CO2 concentration in the field on the quantum yield of photosynthesis of C3 sedge, Scripus olneyi. Plant Physiol 96 221–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long SP, Farage PK, Garcia RL (1996) Measurement of leaf and canopy photosynthetic CO2 exchange in the field. J Exp Bot 47 1629–1642 [Google Scholar]
- Long SP, Zhu XG, Naidu SL, Ort DR (2006) Can improvement in photosynthesis increase crop yields? Plant Cell Environ 29 315–330 [DOI] [PubMed] [Google Scholar]
- Niklas K (1999) Evolutionary walks through a land plant morphospace. J Exp Bot 50 39–52 [Google Scholar]
- Oliver DJ, Raman R (1995) Glycine decarboxylase: protein chemistry and molecular biology of the major protein in leaf mitochondria. J Bioenerg Biomembr 27 407–414 [DOI] [PubMed] [Google Scholar]
- Pearcy RW (1983) The light environment and growth of C3 and C4 tree species in the understory of a Hawaiian forest. Oecologia 58 19–25 [DOI] [PubMed] [Google Scholar]
- Pearcy RW (1990) Sunflecks and photosynthesis in plant canopies. Annu Rev Plant Physiol Plant Mol Biol 41 421–453 [Google Scholar]
- Pearcy RW, Gross LJ, He D (1997) An improved dynamic model of photosynthesis for estimation of carbon gain in sunfleck light regimes. Plant Cell Environ 20 411–424 [Google Scholar]
- Pettersson G (1997) Control properties of the Calvin photosynthesis cycle at physiological carbon dioxide concentrations. Biochim Biophys Acta 1322 173–182 [Google Scholar]
- Pettersson G, Ryde-Pettersson U (1988) A mathematical model of the Calvin photosynthesis cycle. Eur J Biochem 175 661–672 [DOI] [PubMed] [Google Scholar]
- Poolman MG (1999) Computer modeling applied to the Calvin cycle. PhD thesis. Oxford Brookes University, Oxford
- Poolman MG, Fell DA, Thomas S (2000) Modelling photosynthesis and its control. J Exp Bot 51 319–328 [DOI] [PubMed] [Google Scholar]
- Poolman MG, Olcer H, Lloyd JC, Raines CA, Fell DA (2001) Computer modelling and experimental evidence for two steady states in the photosynthetic Calvin cycle. Eur J Biochem 268 2810–2816 [DOI] [PubMed] [Google Scholar]
- Portis AR (1995) The regulation of Rubisco by Rubisco activase. J Exp Bot 46 1285–1291 [DOI] [PubMed] [Google Scholar]
- Portis AR (2003) Rubisco activase—Rubisco's catalytic chaperone. Photosynth Res 75 11–27 [DOI] [PubMed] [Google Scholar]
- Prentice IC, Farquhar GD, Fasham MJR, Goulden ML, Heimann M, Jaramillo VJ, Kheshgi HS, Le Quéré C, Scholes RJ, Wallace DWR (2001) The carbon cycle and atmospheric carbon dioxide. In JT Houghton, Y Ding, DJ Griggs, M Noguer, PJ van der Linden, X Dai, K Maskell, C Johnson, eds, Climate Change 2001: The Scientific Basis. Cambridge University Press, Cambridge, UK, pp 183–237
- Price GD, von Caemmerer S, Evans JR, Siebke K, Anderson JM, Badger MR (1998) Photosynthesis is strongly reduced by antisense suppression of chloroplastic cytochrome bf complex in transgenic tobacco. Aust J Plant Physiol 25 445–452 [Google Scholar]
- Pyke KA, Leech RM (1987) The control of chloroplast number in wheat mesophyll cells. Planta 170 416–420 [DOI] [PubMed] [Google Scholar]
- Raines CA (2003) The Calvin cycle revisited. Photosynth Res 75 1–10 [DOI] [PubMed] [Google Scholar]
- Raines CA, Harrison EP, Olcer H, Lloyd JC (2000) Investigating the role of the thiol-regulated enzyme sedoheptulose-1,7-bisphosphatase in the control of photosynthesis. Physiol Plant 110 303–308 [Google Scholar]
- Raines CA, Lloyd JC, Dyer TA (1999) New insights into the structure and function of sedoheptulose-1,7-bisphosphatase; an important but neglected Calvin cycle enzyme. J Exp Bot 50 1–8 [Google Scholar]
- Schimkat D, Heineke D, Heldt HW (1990) Regulation of sedoheptulose-1,7-bisphosphatase by sedoheptulose-7-phosphate and glycerate, and of fructose-1,6-bisphosphatase by glycerate in spinach chloroplasts. Planta 181 97–103 [DOI] [PubMed] [Google Scholar]
- Segel IH (1975) Enzyme Kinetics. John Wiley & Sons, New York
- Sharkey TD (1985) O2 insensitive photosynthesis in C3 plants. Plant Physiol 78 71–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schurmann P, Jacquot JP (2000) Plant thioredoxin systems revisited. Annu Rev Plant Physiol Plant Mol Biol 51 371–400 [DOI] [PubMed] [Google Scholar]
- Stitt M, Quick WP, Schurr U, Schulze ED, Rodermel SR, Bogorad L (1991) Decreased ribulose-1,5-bisphosphate carboxylase-oxygenase in transgenic tobacco transformed with antisense rbcs. 2. Flux control coefficients for photosynthesis in varying light, CO2, and air humidity. Planta 183 555–566 [DOI] [PubMed] [Google Scholar]
- Tamoi M, Nagaoka M, Miyagawa Y, Shigeoka S (2006) Contribution of fructose-1,6-bisphosphatase and sedoheptulose-1,7-bisphosphatase to the photosynthetic rate and carbon flow in the Calvin cycle in transgenic plants. Plant Cell Physiol 47 380–390 [DOI] [PubMed] [Google Scholar]
- von Caemmerer S (2000) Biochemical Models of Leaf Photosynthesis. Techniques in Plant Sciences Series. CSIRO Publishing, Victoria, Australia, pp 1–28
- Walker D (1992) Concerning oscillations. Photosynth Res 34 387–395 [DOI] [PubMed] [Google Scholar]
- Wingler A, Ann VJ, Lea PJ, Leegood RC (1999) Serine:glyoxylate aminotransferase exerts no control on photosynthesis. J Exp Bot 50 719–722 [Google Scholar]
- Wingler A, Lea PJ, Leegood RC (1997) Control of photosynthesis in barley plants with reduced activities of glycine decarboxylase. Planta 202 171–178 [Google Scholar]
- Winter H, Robinson DG, Heldt HW (1993) Subcellular volumes and metabolite concentrations in barley leaves. Planta 191 180–190 [Google Scholar]
- Winter H, Robinson DG, Heldt HW (1994) Subcellular volumes and metabolite concentrations in spinach leaves. Planta 193 530–535 [Google Scholar]
- Wong S-C, Cowan IR, Farquhar GD (1979) Stomatal conductance correlates with photosynthetic capacity. Nature 282 424–426 [Google Scholar]
- Woodrow IE, Berry JA (1988) Enzymatic regulation of photosynthetic CO2 fixation in C3 plants. Annu Rev Plant Physiol Plant Mol Biol 39 533–594 [Google Scholar]
- Woodrow IE, Mott KA (1993) Modeling C3 photosynthesis—a sensitivity analysis of the photosynthetic carbon reduction cycle. Planta 191 421–432 [Google Scholar]
- Wullschleger SD (1993) Biochemical limitations to carbon assimilation in C3 plants: a retrospective analysis of the A/Ci curves from 109 species. J Exp Bot 44 907–920 [Google Scholar]
- Zhu X-G, Ort DR, Whitmarsh J, Long SP (2004. a) The slow reversibility of photosystem II thermal energy dissipation on transfer from high to low light may cause large losses in carbon gain by crop canopies. A theoretical analysis. J Exp Bot 55 1167–1175 [DOI] [PubMed] [Google Scholar]
- Zhu X-G, Portis AR Jr, Long SP (2004. b) Would transformation of C3 crop plants with foreign Rubisco increase productivity? A computational analysis extrapolating from kinetic properties to canopy photosynthesis. Plant Cell Environ 27 155–165 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.