Abstract
Microtubule organization is intimately associated with cellulose microfibril deposition, central to plant secondary cell wall development. We have determined that a relatively large suite of eight α-TUBULIN (TUA) and 20 β-TUBULIN (TUB) genes is expressed in the woody perennial Populus. A number of features, including gene number, α:β gene representation, amino acid changes at the C terminus, and transcript abundance in wood-forming tissue, distinguish the Populus tubulin suite from that of Arabidopsis thaliana. Five of the eight Populus TUAs are unusual in that they contain a C-terminal methionine, glutamic acid, or glutamine, instead of the more typical, and potentially regulatory, C-terminal tyrosine. Both C-terminal Y-type (TUA1) and M-type (TUA5) TUAs were highly expressed in wood-forming tissues and pollen, while the Y-type TUA6 and TUA8 were abundant only in pollen. Transcripts of the disproportionately expanded TUB family were present at comparatively low levels, with phylogenetically distinct classes predominating in xylem and pollen. When tension wood induction was used as a model system to examine changes in tubulin gene expression under conditions of augmented cellulose deposition, xylem-abundant TUA and TUB genes were up-regulated. Immunolocalization of TUA and TUB in xylem and phloem fibers of stems further supported the notion of heavy microtubule involvement during cellulose microfibril deposition in secondary walls. The high degree of sequence diversity, differential expansion, and differential regulation of Populus TUA and TUB families may confer flexibility in cell wall formation that is of adaptive significance to the woody perennial growth habit.
Polymers of the α-TUBULIN (TUA) and β-TUBULIN (TUB) proteins comprise dynamic arrays of cortical microtubules (MTs) that are continually reorganizing in response to developmental and environmental cues (e.g. Wasteneys, 2004), and are postulated to guide the deposition of cellulose microfibrils during cell wall formation in plants (Ledbetter and Porter, 1963). TUA and TUB proteins exhibit strong sequence conservation, with animal, plant, protist, and fungal isoforms typically sharing >88% amino acid sequence similarity (Fosket and Morejohn, 1992; Dutcher, 2001). Functional conservation of tubulins across kingdoms is supported by copolymerization of heterologous or chimeric TUA and TUB, either in vitro (Bondstar et al., 1986) or in vivo (Anthony and Hussey, 1999; Anthony et al., 1999). However, plant and animal tubulins differ in their sensitivities to various anti-MT drugs (Morejohn and Fosket, 1984), in accordance with plant- and vertebrate-specific TUA and TUB amino acid residues at consensus positions (Fosket and Morejohn, 1992). Functional heterogeneity of tubulin subunits within species, organs, or even cells is further manifest as spatiotemporally distinct gene products, many of which are subject to posttranslational modification (PTM; for review, see Luduena, 1998; McKean et al., 2001).
Spatiotemporal control of tubulin gene expression has been documented in all plant species examined. For instance, Arabidopsis (Arabidopsis thaliana) contains six TUAs encoding four distinct proteins (Kopczak et al., 1992). ArathTUA1 is expressed specifically in pollen (Carpenter et al., 1992), while the other five are expressed in vegetative tissues (Kopczak et al., 1992; Abe et al., 2004). Of the nine Arabidopsis TUBs, ArathTUB1 and ArathTUB5 are preferentially expressed in roots and leaves, respectively, while ArathTUB9 accumulates specifically in pollen (Snustad et al., 1992; Cheng et al., 2001). Rice (Oryza sativa) also contains a pollen-specific isoform (OryzaTUB8) and seven other TUBs that show variable expression during development (Yoshikawa et al., 2003). Strict temporal association of specific tubulin transcripts during cotton (Gossypium hirsutum) fiber development (Whittaker and Triplett, 1999; Li et al., 2002) or Zinnia tracheary element differentiation (Yoshimura et al., 1996) is consistent with the idea of a functional link between MTs and microfibril deposition during secondary wall synthesis. The five characterized TUAs of cotton are highly expressed in elongating fibers, but only GoshiTUA2/3 and GoshiTUA4 remain abundant following the onset of secondary cell wall synthesis (Whittaker and Triplett, 1999). In Zinnia, transcripts of ZinelTUB1 and ZinelTUB3 increase during trans-differentiation of mesophyll cells into tracheary elements, contrasting with the constitutive and weak expression of ZinelTUB2 throughout the culture period (Yoshimura et al., 1996).
These data suggest a basis for examining the role of tubulin isoforms during secondary wall development in wood-forming tissues of trees, a process characterized by orderly deposition of extensive microfibril arrays. Changes in the alignment of cellulose microfibrils in developing cell walls of xylem and phloem have in fact been linked to changes in MT orientation in both hardwood and softwood tree species (Abe et al., 1995; Chaffey et al., 1997, 1999). A recent report on functional associations between cellulose synthase complexes and MTs by time-lapse visualization provides further evidence for a direct cytoskeletal guidance mechanism during cellulose deposition (Paredez et al., 2006). With the availability of the sequenced Populus genome, an in-depth analysis of the role of tubulin gene expression in wood-forming tissues can be initiated. We report here that the TUA and TUB gene families are differentially expanded in Populus compared to their herbaceous counterparts. Their encoded protein sequences are more diverse than in any other species reported so far, especially at the hypervariable C terminus. Both the TUA and TUB families contain spatiotemporally distinct isoforms, a small minority of which are highly expressed in wood-forming tissues undergoing secondary cell wall thickening. The abundance and multiplicity of xylem-expressed TUA and TUB isoforms certainly distinguish “wood” formation in Populus and Arabidopsis. Over the course of tree ontogeny, such flexibility of expression might reasonably be expected to contribute adaptively to perennial cellulose deposition and vascular development.
RESULTS
Cloning of Aspen TUA Genes
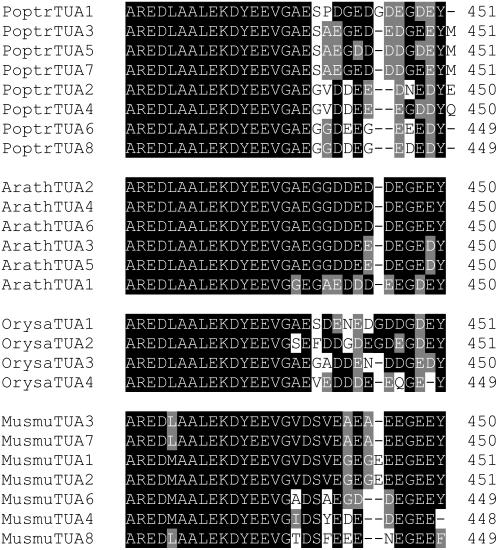
Eight TUA cDNAs, designated TUA1 through TUA8, were isolated from quaking aspen (Populus tremuloides Michx.). TUA1 was originally identified as a xylem-abundant (relative to phloem) cDNA fragment by mRNA differential display (Touchell et al., 2003) and its full-length cDNA subsequently isolated by screening an aspen xylem cDNA library. TUA2 to TUA8 were cloned from various aspen tissues using PCR-based approaches (see “Materials and Methods”). The assembly v1.1 of the recently released Populus trichocarpa genome (Tuskan et al., 2006) contains all eight TUA orthologs (annotated as PoptrTUA1 through PoptrTUA8), with two additional truncated sequences, PoptrTUA5tψ and PoptrTUA7tψ (Table I), which show highest nucleotide identity to PoptrTUA5 and PoptrTUA7, respectively, and are most likely pseudogenes. The open reading frames of the eight TUA genes range from 1,350 to 1,356 bp in size, and share 74% to 97% sequence identity. However, they contain distinct 3′-untranslated regions (UTRs), with sequence identity less than 62%. The eight predicted TUA proteins range in length from 449 to 551 amino acids, and show a very high degree of sequence homology to each other (88%–98% identity; Supplemental Fig. S1), except for their hypervariable C termini (Fig. 1). All plant TUA proteins identified so far contain a C-terminal Tyr residue (referred to as Y-type), which has been implicated in PTM control of tubulin stability in animal models. Interestingly, only three of the eight predicted aspen TUA proteins are Y-type. The other five terminate with either Met (i.e. M-type: class I TUA3, TUA5, and TUA7) or Glu/Gln (i.e. E/Q-type: class II TUA2 and TUA4; Fig. 1).
Table I.
List of TUA and TUB genes identified from the Populus genome
| Gene | JGI Gene Model | Locus |
|---|---|---|
| PoptrTUA1 | gw1.II.3483.1 | LG_II:8299090–8300967 |
| PoptrTUA2 | estExt_fgenesh4_pm.C_LG_III0736 | LG_III:18595875–18598862 |
| PoptrTUA3 | estExt_Genewise1_v1.C_LG_I4174 | LG_I:27533386–27535822 |
| PoptrTUA4 | estExt_fgenesh4_pg.C_LG_I0041 | LG_I:292815–295515 |
| PoptrTUA5 | eugene3.00090803 | LG_IX:4925341–4927829 |
| PoptrTUA6 | eugene3.00130559 | LG_XIII:4169981–4173491 |
| PoptrTUA7 | estExt_Genewise1_v1.C_LG_XVII1003 | LG_XVII:4138330–4140210 |
| PoptrTUA8 | estExt_fgenesh4_pg.C_LG_XIX0417 | LG_XIX:5204194–5207599 |
| PoptrTUA5tψ | gw1.III.2686.1 | LG_III:12748887–12749356 |
| PoptrTUA7tψ | gw1.VII.1672.1 | LG_VII:3417187–3417348 |
| PoptrTUB1 | eugene3.00011551 | LG_I:17121298–17123342 |
| PoptrTUB2 | estExt_fgenesh4_pm.C_LG_IX0574 | LG_IX:8108706–8110629 |
| PoptrTUB3 | eugene3.00010923 | LG_I:7896652–7899456 |
| PoptrTUB4 | eugene3.00030977 | LG_III:11846773–11849592 |
| PoptrTUB5 | fgenesh4_pg.C_LG_VI000763 | LG_VI:6461827–6463345 |
| PoptrTUB6 | eugene3.00161038 | LG_XVI:10159508–10161158 |
| PoptrTUB7 | estExt_Genewise1_v1.C_LG_I6025 | LG_I:34930733–34933465 |
| PoptrTUB8 | eugene3.01400079 | scaffold_140:531237–534474 |
| PoptrTUB9 | eugene3.00010909 | LG_I:7810326–7812139 |
| PoptrTUB10 | grail3.0018029802 | LG_III:11963516–11965115 |
| PoptrTUB11 | eugene3.00120398 | LG_XII:3902075–3904191 |
| PoptrTUB12 | estExt_fgenesh4_pm.C_LG_XV0128 | LG_XV:3923544–3925244 |
| PoptrTUB13 | eugene3.00011808 | LG_I:19840697–19843574 |
| PoptrTUB14 | estExt_fgenesh4_pm.C_LG_IX0458 | LG_IX:6223860–6226800 |
| PoptrTUB15 | estExt_Genewise1_v1.C_LG_I1970 | LG_I:19849822–19852686 |
| PoptrTUB16 | estExt_fgenesh4_pm.C_LG_IX0457 | LG_IX:6218943–6221998 |
| PoptrTUB17 | eugene3.00060314 | LG_VI:2140954–2144202 |
| PoptrTUB18 | eugene3.00160267 | LG_XVI:1699400–1702625 |
| PoptrTUB19 | estExt_Genewise1_v1.C_LG_II0459 | LG_II:1312352–1316028 |
| PoptrTUB20 | grail3.0002068402 | LG_V:16706215–16709216 |
| PoptrTUB8tψ | eugene3.00121178 | LG_XII:13261307–13261577 |
| PoptrTUB18tψ | grail3.0047019501 | LG_III:15713547–15713716 |
Figure 1.
C-terminal sequence alignments of TUA families from Populus, Arabidopsis, rice, and mouse. GenBank accession numbers of the Arabidopsis, rice, and mouse proteins are provided in Supplemental Table S2.
Phylogenetic Analysis of the TUA Family
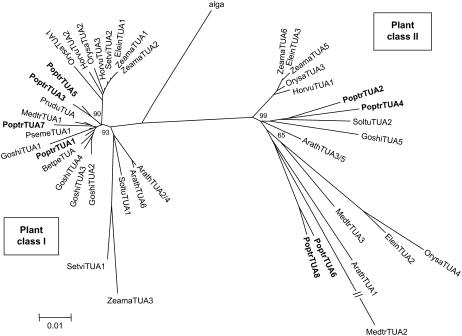
Phylogenetic analysis of representative algal and plant full-length TUA proteins is shown in Figure 2. Plant TUAs form two distinct classes, with the eight Populus, six Arabidopsis, and four rice isoforms evenly distributed between the two classes. Although the bootstrap values for most nodes were low, likely due to high degree of sequence similarity, a similar topology was observed using the neighbor-joining or minimum-evolution methods with interior-branch test (only the minimum-evolution tree is shown). In general, monocot and dicot TUAs cluster separately within a class (Fig. 2). In class I, the xylem-originated PoptrTUA1 clusters with fiber-specific GoshiTUA2, GoshiTUA3, and GoshiTUA4 from cotton, consistent with involvement in secondary cell wall development. However, the three Arabidopsis class I members cluster closely with potato (Solanum tuberosum; SoltuTUA1, ABB02631) and two monocot TUAs from maize (Zea mays; ZeamaTUA3, CAA44861) and green foxtail (Setaria viridis; SetviTUA1, CAE52514) in a separate branch. Similarly, the class II family also includes a third and distinct branch containing both dicot and monocot TUA members, one of which (ArathTUA1) was shown to exhibit pollen-specific expression (Carpenter et al., 1992).
Figure 2.
Minimum-evolution tree of representative full-length plant TUA proteins. The green algal (Chlamydomonas reinhardtii) TUA (AAN87017) was used as the outgroup. Numbers on major branches indicate percentage support of interior-branch test using 1,000 replicates. GenBank accession numbers of the sequences included are provided in Supplemental Table S2.
The four Populus class II TUAs probably descend from one ancient gene that gave rise to the progenitors of PoptrTUA2/4 and PoptrTUA6/8 during the eurosid genome-wide duplication, and eventually to the modern complement of PoptrTUA2, PoptrTUA4, PoptrTUA6, and PoptrTUA8 following the salicoid duplication (Tuskan et al., 2006). This duplication was dated between 6 and 10 million years ago (mya), based on synonymous substitution rates of 1.5 × 10−8 and 9.1 × 10−9 mutations per site per year for dicot genes by Koch et al. (2000) and Lynch and Conery (2000), respectively. Two of the class II Arabidopsis members, ArathTUA3 and ArathTUA5, are tandem repeats encoding identical proteins. This duplication was estimated to have occurred only 3 to 5 mya. None of the other Populus and Arabidopsis TUA genes is associated with genome-wide or tandem duplications, suggesting that they may have originated from other lineage-specific segmental gene duplication events. For instance, PoptrTUA5 and PoptrTUA7 seem to have duplicated independently around the same time as the recent Populus whole-genome duplication event, followed by truncations of PoptrTUA5tψ and PoptrTUA7tψ. ArathTUA2 and ArathTUA4 also encode identical proteins, and appear to have duplicated as an independent event 5 to 8.5 mya.
Gene Structure of the TUA Family
TUA genomic sequences were retrieved from the Populus genome portal v1.1 for gene structure analysis. The exon-intron splice junctions of all eight full-length TUA genes follow the GT-AG rule, but class I and class II PoptrTUAs differ both in number and position of introns (Fig. 3A). Three introns are located within the coding region of the class I members, whereas four are present in class II genes. While all intron positions are conserved within each subfamily, only the second intron position is common to all PoptrTUAs (Fig. 3A). Analysis of available genomic sequences from public databases also showed a conserved exon-intron structure among the class II Arabidopsis, maize, and rice TUA genes, but this is not the case for the class I family. Of the three Arabidopsis class I members, only ArathTUA2 exhibits conserved position of all three introns. The positions of introns 1 and 3 are conserved in ArathTUA4, whereas only the position of the first intron is conserved in ArathTUA6 (Kopczak et al., 1992). Other class I TUA genes from maize (Montoliu et al., 1990), rice (The Institute for Genomic Research Rice Database), green foxtail (Delye et al., 2004), and pea (Pisum sativum; Brierley et al., 1995) all contain three introns. Taken together, the distinct phylogenetic relationship and gene structures among class I and class II TUA members suggest their divergence before the separation of monocot and dicot plants.
Figure 3.
Populus TUA and TUB gene structure. A, Class I TUAs contain three introns, while class II TUAs have four. Intron positions are conserved within each class, but only the second intron is conserved in all TUA genes (dashed lines denote position conservation). B, All TUB genes have two introns within their coding sequences at conserved positions. Class I members contain an additional intron in the 5′-UTR. Class designation is marked by vertical lines on the left, with a dashed line representing the class I-like TUB group.
In Silico Identification of Populus TUB Genes
Twenty TUB genes, designated PoptrTUB1 through PoptrTUB20, were identified from the P. trichocarpa genome sequence and confirmed to be transcriptionally active in aspen (see below). The manually curated gene models are listed in Table I. The predicted open reading frames range in size from 1,335 to 1,356 bp, and share 76% to 96% nucleotide identity. Their 3′-UTRs are more variable, exhibiting less than 68% sequence identity (not shown). The hypothetical PoptrTUB proteins vary from 444 to 451 amino acids in length, and, like PoptrTUAs, are highly conserved (89%–98% sequence identity overall; Supplemental Fig. S2), except for the hypervariable C termini (Fig. 4). Compared to the PoptrTUA family, the PoptrTUB family has undergone significant expansion, involving both eurosid and salicoid genome-wide duplication events, along with tandem gene duplication, giving rise to 10 pairs of highly homologous PoptrTUB genes. The tandem gene duplication event involving PoptrTUB13 and PoptrTUB15 is estimated to have occurred 4 to 7 mya, and the second event involving PoptrTUB14 and PoptrTUB16 6 to 10.5 mya.
Figure 4.
C-terminal sequence alignments of TUB families from Populus, Arabidopsis, rice, and mouse. GenBank accession numbers of the Arabidopsis, rice, and mouse proteins are provided in Supplemental Table S2.
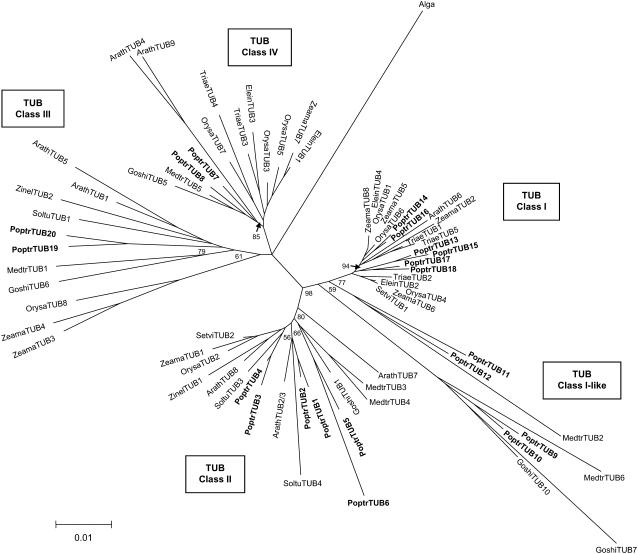
Phylogenetic analysis reveals at least four distinct classes of plant TUBs (Fig. 5). Class I and class I-like comprise by far the largest cluster, containing half of the Populus (10 out of 20) and known maize (four out of eight) TUB families, along with a single Arabidopsis member (ArathTUB6). Four TUBs (PoptrTUB9–PoptrTUB12) are designated as class I-like, as they lack a 5′-UTR intron that is characteristic of class I members (Fig. 3B) and do not form a tight cluster with class I TUBs (Fig. 5). Class II consists primarily of dicot TUBs, including six Populus and four Arabidopsis isoforms, and one each from rice, maize, and green foxtail. Class III and Class IV each contain two Populus and two Arabidopsis isoforms arising from genome-wide duplications. Class III members are unique in that many contain an insertion of one to three amino acids at position 39, and others have amino acid hypervariability at or near this position. Several class III and class IV members, including ArathTUB9 (Cheng et al., 2001), OrysaTUB8 (Yoshikawa et al., 2003), ZeamaTUB3, and ZeamaTUB4 (Rogers et al., 1993), were found to be preferentially expressed in pollen. The Populus genome also contains two truncated TUB pseudogenes, designated PoptrTUB8tψ and PoptrTUB18tψ based on their closest homologs, PoptrTUB8 and PoptrTUB18, respectively. The PoptrTUB18-PoptrTUB18tψ duplication probably occurred around the salicoid genome duplication, while the PoptrTUB8-PoptrTUB8tψ duplication was more recent, dated 3 to 5 mya based on molecular clock analysis.
Figure 5.
Minimum-evolution tree of representative full-length plant TUB proteins. The green algal (C. reinhardtii) TUB (P04690) was used as the outgroup. Numbers on major branches indicate percentage support of interior-branch test using 1,000 replicates. GenBank accession numbers of the sequences included are provided in Supplemental Table S2.
Gene Structure of the TUB Family
All PoptrTUB genes have two introns located at conserved positions (Fig. 3B), analogous to the gene structure of the Arabidopsis TUB family previously reported (Snustad et al., 1992). This intron structure is also observed in 12 other plant TUB genes for which genomic sequences are available (data not shown). The only known exceptions are class II maize ZeamaTUB1 and rice OrysaTUB2, both of which contain only the first intron, while the closely related SetviTUB2 from green foxtail has two. This suggests that the second intron was lost independently in two different lineages. The first event occurred after the ancestral divergence of rice and maize/green foxtail in the lineage leading to rice. The second event occurred after the ancestral divergence of maize and green foxtail in the lineage leading to maize. Class I TUBs, such as PoptrTUB13 to PoptrTUB18 (Fig. 3B), ArathTUB6, SetviTUB1, ZeamaTUB2, ZeamaTUB5, OrysaTUB4, and OrysaTUB6, contain an additional intron within the 5′-UTR. In the case of OrysaTUB4, this leader intron has been shown to positively regulate transcription both in vivo and in vitro (Morello et al., 2002). Taken together, the presence of four distinct classes with members from both dicot and monocot species suggests that the angiosperm TUB family members may have originated from a single ancestral gene.
Expression Patterns of TUA and TUB Transcripts
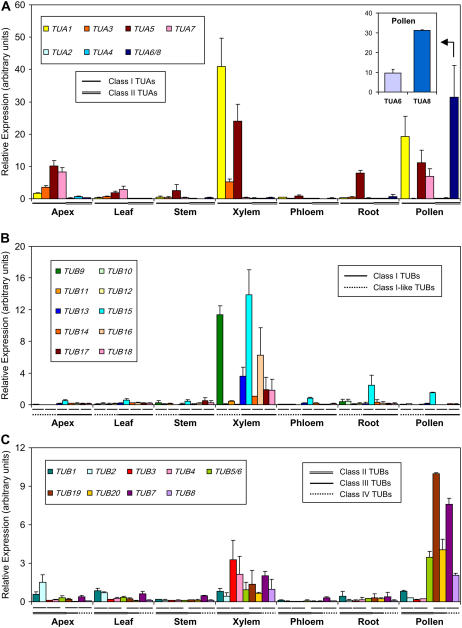
Expression of the eight TUA and 20 TUB genes in various aspen tissues was analyzed by quantitative real-time reverse transcription (RT)-PCR using gene-specific primers (Supplemental Table S1). The class I TUA members, with the exception of TUA7, were most abundantly expressed in developing xylem of field-grown trees. The Y-type TUA1 was by far the most abundant transcript in developing xylem, consistent with its origin as a xylem-specific cDNA, followed by the M-type TUA5 (Fig. 6A). TUA5 was also highly expressed in root tips. Transcripts of the other two M-type class I isoforms were detected at a lower level, with TUA3 primarily in xylem and shoot apices, and TUA7 in apices and young leaves. It appears that both Y- and M-type class I members were abundant in xylem, whereas M-type class I TUAs predominated in all other vegetative tissues. Expression of class II TUAs was very weak, with no apparent preference among vegetative tissues. Aspen pollen grains contained a high level of tubulin transcripts, including both Y- (TUA1) and M-type (TUA5 and TUA7) class I TUA members, and, most notably, the Y-type class II TUA6/8 that were negligibly expressed elsewhere (Fig. 6A). Not surprisingly, the predicted TUA6 and TUA8 proteins cluster closely with the pollen-specific ArathTUA1 in phylogenetic analysis (Fig. 2), suggesting functional conservation. Because of the high degree of sequence identity, RT-PCR primers were initially designed to amplify both TUA6 and TUA8, and detection of both transcripts was then confirmed by sequencing of randomly selected RT-PCR clones (data not shown). Additional gene-specific primers were designed to distinguish TUA6 and TUA8 expression in pollen. Real-time RT-PCR analysis showed that both genes were well expressed in pollen, but TUA8 transcripts were 3-fold more abundant than TUA6 (Fig. 6A, inset).
Figure 6.
Relative TUA and TUB transcript levels in various aspen tissues. Expression levels were normalized to the geometric mean of three housekeeping genes. Error bars represent the measurement range of two biological replicates. A, Relative expression of TUAs. Both TUA6 and TUA8 transcripts were detected by the TUA6/8 primers. Inset, Gene-specific detection of TUA6 and TUA8 transcript levels in pollen. B and C, Relative expression of TUBs. Both TUB5 and TUB6 transcripts were detected by the TUB5/6 primers. Different TUA and TUB classes are denoted below the x axis by solid, double, or dashed lines as shown in the legend of each section. Hairlines immediately below the x axis in B and C specify paralogous TUB gene pairs. Primer sequences are provided in Supplemental Table S1.
TUB9 and TUB15 represent the predominant TUB species in developing xylem, followed by TUB16 and TUB13 (Fig. 6). Transcripts of TUB15 were also detected in roots and pollen grains. The other TUB genes were expressed at low levels in vegetative tissues, but many of the class II, III, and IV TUB members were well expressed in pollen (Fig. 6C). Pollen expression of class III TUB19 and TUB20 and class IV TUB7 and TUB8 is consistent with their phylogenetic clustering with other pollen-specific TUB isoforms from Arabidopsis, rice, and maize (Fig. 5). We noticed that expression of TUB paralogs, such as class I TUB9 versus TUB10, TUB13 versus TUB15, and TUB14 versus TUB16, differed, consistent with the possibility of functional divergence. This has also been noted in Arabidopsis for its class III members, with ArathTUB1 exhibiting root-preferential expression and ArathTUB5 being leaf abundant (Snustad et al., 1992).
Expression of TUA and TUB Transcripts in Tension Wood
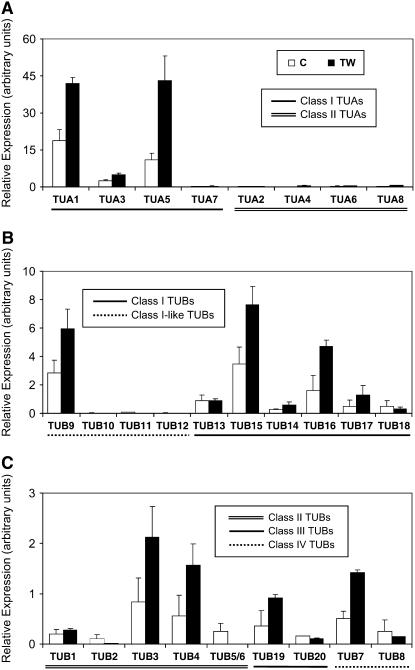
Tension wood is an angiosperm-specific response to gravitational stress, characterized by altered microfibril structure and properties due to the presence of a gelatinous layer that is composed primarily of cellulose (Norberg and Meier, 1966). An increased abundance of MTs, in parallel with microfibrils, has been noted in tension wood fibers (Fujita et al., 1974; Prodhan et al., 1995). We therefore measured TUA and TUB expression in tension wood tissue. As shown in Figure 7, xylem-abundant TUA1 and TUA5 were up-regulated 2- to 4-fold in tension wood. Transcripts of weakly expressed class II TUA4 and TUA8 were also induced. Similarly, expression of xylem-preferential TUB9, TUB15, and TUB16 was up-regulated by tension stress, as was the less abundant TUB3, TUB4, TUB7, and TUB19. Similar up-regulation of three secondary cell wall-associated cellulose synthase genes and one Korrigan endoglucanase in experimentally induced tension wood has also been reported in aspen (Bhandari et al., 2006). Thus, xylem-abundant TUA and TUB genes are coordinately regulated with cellulose biosynthesis machinery.
Figure 7.
Relative transcript levels of TUA and TUB genes in developing xylem of normal (C) and tension wood (TW) tissues from aspen. Expression levels were normalized to the geometric mean of three housekeeping genes. Error bars represent the measurement range of two biological replicates.
Fiber-Specific Localization of Tubulins in Developing Stems
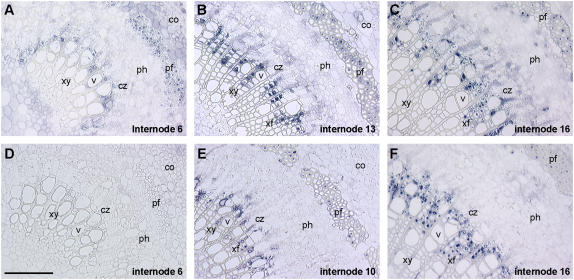
Cellular distribution of tubulin proteins in aspen stem sections was investigated by immunolocalization (Fig. 8). Using a polyclonal anti-TUA antibody raised against recombinant aspen TUA1, no specific signal was detected at the third internode (data not shown), but conspicuous staining in developing phloem fibers and newly formed xylem fibers was observed at the sixth internode (Fig. 8A). The signal intensified in stem sections undergoing secondary cell wall thickening (Fig. 8, B, C, E and F). In xylem, the strongest signal was found in developing fibers, with weak staining observed in some ray parenchyma and newly developed vessel cells adjacent to the cambial zone. Strong signals were also found in phloem fibers with extensive secondary cell wall development. Weak signals were observed in ray initials of the cambial zone and in some newly developed phloem cells. Similar immunolocalization patterns were observed using a commercially available polyclonal anti-TUB peptide-antibody (abcam ab15568; Fig. 8, E and F). The weaker TUB signals from thin-walled ray parenchyma, phloem, and pith cells (Fig. 8, E and F versus B and C) may reflect differential sensitivity of single-epitope (TUB) versus purified recombinant fusion protein (TUA) antigens. Although we do not ascribe the anti-TUA and anti-TUB signals to specific tubulin isoforms, the results show nevertheless that the most abundant tubulin species in wood-forming tissues are localized to fiber cells.
Figure 8.
TUA and TUB protein localization in aspen stem sections. The most abundant TUA and TUB immunosignals are detected in xylem and phloem fibers of developing stem sections, using polyclonal antibodies raised against recombinant aspen TUA1 (A–C) or TUB peptide (E and F). A preimmune serum was used as control (D). Scale bar = 100 μm. co, Cortex; cz, cambial zone; pf, phloem fiber; ph, phloem; v, vessel; xf, xylem fiber; xy, xylem.
DISCUSSION
In mammalian systems (e.g. human and mouse), the TUA and TUB families are identical in size, with seven genes each (Sullivan, 1988; Stanchi et al., 2000). The family sizes, however, vary in higher plants, with six TUA and nine TUB genes in Arabidopsis, expanding to eight TUAs and 20 TUBs in the recently sequenced woody perennial Populus. We now know that Populus experienced two genome-wide gene duplication (eurosid and salicoid) events, followed by a series of chromosomal reorganizations, involving reciprocal tandem/terminal fusions and translocations (Tuskan et al., 2006). Interestingly, all 20 PoptrTUB genes are associated with genome-wide or tandem gene duplication events, whereas only four of the eight PoptrTUA genes are so derived. What led to the distinct patterns of gene retention (with regard to TUB) and/or loss (with regard to TUA) for two families of proteins that form 1:1 heterodimers is not known. Quantitative RT-PCR detection and sequence confirmation of all 20 TUB amplicons in various aspen tissues excludes the possibility that any TUB gene is not expressed.
When viewed in the context of coordinated regulation of TUA and TUB in other species, the selective expansion of the TUB gene family in Populus is interesting. In yeast (Saccharomyces cerevisiae), overexpression of TUA or TUB can lead to arrest of cell division, but a lethal phenotype was observed only in TUB-overexpressing strains, suggesting that transcript levels of TUB in excess of TUA are uniquely toxic (Weinstein and Solomon, 1990). Cotransformation of TUA and TUB was an absolute requirement for recovery of transgenic maize and tobacco (Nicotiana tabacum), as overexpression of either TUA or TUB alone was lethal (Anthony and Hussey, 1998, 1999). In the case of aspen, attempts to transgenically manipulate TUA1 expression also failed to produce viable transformants via Agrobacterium transformation and organogenesis (R.V. Oakley and C.-J. Tsai, unpublished data), suggesting that tight regulation of the TUA:TUB transcript ratio is also important in Populus. Indeed, real-time RT-PCR analysis suggests that compensatory expression may be important to maintain a viable functional balance of TUA:TUB in Populus. In xylem, for instance, fewer TUA (e.g. TUA1 and TUA5) than TUB (e.g. TUB9, TUB13, TUB15, and TUB16) genes were expressed, but the relative transcript levels of the two xylem-predominant TUAs were much higher than those of the TUBs. The same principle appeared to operate in pollen. The finding that xylem-abundant TUA, but not TUB, isoforms are also highly expressed in pollen is interesting, and may be viewed as a possible outcome of the differential evolution of these two gene families in Populus. The smaller TUA family appears to exhibit broader functionality, in this case, regulating wood formation as well as pollen development, whereas the large TUB family exhibits genewise redundancy and classwise functional specialization. Taken together, the variations in TUA and TUB family size and the quantitative differences in expression of individual gene family members lead us to hypothesize that transcriptional regulation is an important aspect of MT function in Populus.
The identification of five novel Populus TUAs with a unique C-terminal Met, Glu, or Gln residue is of particular interest with regard to tyrosination/detyrosination of TUA, a PTM that has been extensively characterized in mammalian systems (MacRae, 1997). It involves the cyclic removal and re-attachment of the C-terminal Tyr (or Phe in MmTUA8; Fig. 1) by tubulin-specific carboxypeptidase and tubulin Tyr ligase (TTL), respectively (Idriss, 2000). Detyrosinated TUA can give rise to Δ2-TUA (also called nontyrosinatable TUA), which is excluded from the tyrosination/detyrosination cycle due to loss of the C-terminal TTL recognition motif GEE(Y) (Rudiger et al., 1994). Interestingly, the TTL motif is poorly conserved in Arabidopsis, rice, and Populus isoforms (Fig. 1). This suggests that detyrosination of plant TUAs, if it occurs, may not be reversible. Consistent with this is the lack of a clear TTL homolog in plants. The single-copy Arabidopsis (At1g77550) and Populus (Joint Genome Institute [JGI] gene model eugene3.00280021) genes annotated as “TTL family protein” encode polypeptides of 855 to 869 amino acids that are much larger than the well-characterized mammalian TTL of 370 amino acids (Ersfeld et al., 1993). Although detyrosinated and nontyrosinatable TUAs have been reported in tobacco suspension cells using mammalian PTM-specific antibodies (Smertenko et al., 1997), they have not been detected in planta (Duckett and Lloyd, 1994; Gilmer et al., 1999). PTMs afford a means to modulate tubulin diversity and MT dynamics in animals (for review, see MacRae, 1997; Westermann and Weber, 2003). Given the greater sequence diversity at the C terminus of Populus TUA and TUB isoforms, it would be of interest to investigate the role and extent of tubulin PTMs in this woody perennial. Such investigation will likely depend on proteomics approaches and development of plant tubulin PTM-specific antibodies.
Our results showed that both TUA and TUB families in Populus contain distinct isoforms, some with particularly strong expression in tissues undergoing secondary cell wall thickening and others with strong expression in pollen. Pollen-specific tubulin isoforms are phylogenetically conserved (Figs. 2 and 5), although the role of MTs in pollen development remains elusive. Recent characterization of pollen-specific kinesin-related motor proteins (Cai et al., 2000; Romagnoli et al., 2003) and MT-associated protein (Huang et al., 2007) support MT's participation with actin filaments in directing vesicle and organelle transport in elongating pollen tubes. MTs likely play a role in deposition of pollen cell wall components, e.g. pectins, callose, and cellulose, and in maintenance of the cylindrical shape of the pollen tube (Cai et al., 2005).
Several lines of evidence suggest that the xylem-predominant TUA and TUB isoforms are specifically associated with cellulose synthesis during secondary cell wall formation. First, TUA and TUB protein immunolocalization signals are strongest in xylem and phloem fiber cells of aspen stems undergoing secondary wall thickening (Fig. 8). Second, several of the xylem-abundant and tension wood-induced TUA and TUB genes were among the most abundant transcripts detected by microarray or EST analysis during normal and tension wood formation in Populus (Hertzberg et al., 2001; Pilate et al., 2004; Andersson-Gunneras et al., 2006). Third, allelic variation in a loblolly pine (Pinus taeda) TUA gene was found to be significantly associated with earlywood microfibril angle in an association genetics study of wood quality traits (Gonzalez-Martinez et al., 2007). The deduced protein sequence of this pine TUA (EST contig 8045 available at http://biodata.ccgb.umn.edu/nsfpine/contig_dir20/) is identical to PsemeTUA1 from Douglas fir (Pseudotsuga menziesii) and clusters closely with class I Populus TUAs (Fig. 2). Finally, silencing the Eucalyptus homolog of PoptrTUB9 has recently been shown to affect cellulose microfibril angle using induced somatic sector analysis (Spokevicius et al., 2007). Available molecular evidence thus supports in situ and, recently, in vivo visualization studies for a role of cortical MTs in the deposition and orientation of cellulose microfibrils (Ledbetter and Porter, 1963; Giddings and Staehelin, 1991; Paredez et al., 2006). In wood fibers undergoing active secondary cell wall synthesis, the process appears to involve specialized TUA and TUB subunits.
Exactly how MTs exert an influence over microfibril deposition remains unclear, but it likely involves multiple factors, including an assortment of MT-associated proteins (Burk and Ye, 2002; Zhong et al., 2002) and biophysical feedback signals from cellulose microfibrils (Fisher and Cyr, 1998; Himmelspach et al., 2003). Perennial secondary growth is the most prominent of the vegetative growth features distinguishing woody species from herbaceous annuals. Although many aspects of wood formation appear to be conserved between Populus and Arabidopsis, e.g. lignin and cellulose biosynthetic gene networks (Ehlting et al., 2005; Geisler-Lee et al., 2006), the underlying cytoskeletal processes orchestrating cellulose MF deposition and orientation may depend on tubulins in ways that are less conserved. TUA and TUB gene expression has not been associated with secondary growth in Arabidopsis (Ko et al., 2004; Ehlting et al., 2005). The discrepancy in tubulin regulation appears to coincide with differential evolution of the secondary cell wall-associated TUA and TUB isoforms. The Arabidopsis class I TUA members are unusual in that two of them lack the conserved class I gene structure, and that they cluster separately from the main dicot class I branch, including the fiber-specific poplar and cotton TUAs. In the case of TUB, the class I and class I-like group is underrepresented in Arabidopsis, with a lone member. By contrast, Populus possesses a disproportionately large group of class I and I-like TUBs (10 of 20), encompassing all of the xylem- and tension wood-predominant isoforms. Taken together, our data suggest that increased tubulin isoform diversity along with elevated transcript abundance characterize wood and fiber formation in Populus. This may be in accordance with expanded transcriptional and posttranslational regulation of tubulin dynamics to sustain long-term, perennial growth of large woody plants. Future investigations into tubulin expression and possible PTMs in response to the life-long continua of developmental and biotic and abiotic cues trees face may be necessary to shed additional light on the functional significance of the expanded tubulin gene families in Populus.
MATERIALS AND METHODS
Plant Materials
Shoot apices, young leaves, stems (internodes 10–14 from the apex), and root tips (terminal 5 mm) were obtained from greenhouse-grown aspen (Populus tremuloides). Developing secondary xylem and phloem were collected from wild aspen trees on the campus of Michigan Technological University during the peak growing season (July to early August) of 1999 through 2004. Developing tension wood xylem was harvested from the upper side of aspen trees bent at a 30 to 40 degree angle from the vertical axis for 3 months. Reproductive branches were taken from individual wild aspen trees in the vicinity of Houghton in April 2006, and maintained in a greenhouse until flowering. Pollen shed from mature male catkins was collected. Tissues were snap-frozen in liquid nitrogen and stored at −80°C until use.
DNA Cloning and Sequencing
Total RNA was extracted according to Chang et al. (1993) and treated with TURBO DNase (Ambion) to remove contaminating DNA. A partial TUA1 fragment was first identified by mRNA differential display following the protocols of Touchell et al. (2003) and the full-length cDNA obtained by screening of an aspen xylem cDNA library (Ge and Chiang, 1996), according to standard recombinant DNA techniques (Sambrook and Russell, 2001). PCR-based approaches were used to clone other tubulin fragments, using cDNA synthesized with SuperScript II reverse transcriptase and an oligo(dT) primer according to the manufacturer's instructions (Invitrogen). Partial cDNA fragments of the other TUA genes were obtained by RT-PCR of phloem (TUA2, TUA5), apex (TUA3), or xylem (TUA4, TUA6, TUA7, TUA8) cDNA, using degenerate or gene-specific primers (Supplemental Table S1). Full-length cDNA clones were obtained by RT-PCR using gene-specific primers (Supplemental Table S1) or by 5′-RACE using the SMART RACE cDNA amplification kit (CLONTECH). PCR products were purified using the UltraClean PCR clean-up kit (MoBio) and cloned into the pCRII vector (Invitrogen). Positive clones were sequenced fully from both directions using the CEQ dye terminator cycle sequencing quick start kit and the CEQ8000 genetic analysis system (Beckman Coulter).
Sequence Analysis
Full-length amino acid sequences were aligned by ClustalW 1.82 (Chenna et al., 2003) and displayed with BOXSHADE 3.21 (http://www.ch.embnet.org/software/BOX_form.html). Phylogenetic analysis was performed with MEGA 3.1 (Kumar et al., 2004), using the neighbor-joining or minimum-evolution methods. Rooted trees were generated from the interior-branch test with 1,000 iterations, using the equal-input model (heterogeneous patterns) and the complete-deletion option for handling alignment gaps. Similar topologies were obtained when the C-terminal hypervariable regions were excluded from the sequence alignment and phylogenetic analyses (not shown). The accession numbers of sequences included in the phylogenetic analyses are listed in Supplemental Table S2. Genomic sequences corresponding to the eight TUA and 20 TUB genes were retrieved from the Populus genome portal (v1.1) hosted at the JGI and the exon-intron structure displayed using the Gene Structure visualization tool (http://warta.bio.psu.edu/cgi-bin/Tools/StrDraw.pl).
Estimation of Gene Duplication Dates
Protein coding regions of genes were aligned and nucleotide substitution rates were estimated using the distance measures of Nei and Gojobori, and the Jukes-Cantor correction implemented in MEGA 3.1. Synonymous substitution rates were used for calculating divergence and duplication times (T) of genes using k = Ks/2T, where k represents absolute rate of synonymous substitution per site per year for dicots, and Ks is the estimated number of synonymous substitutions per site between homologous sequences using the Kimura two-parameter method as described by Gaut et al. (1996) and Ramakrishna et al. (2002). This estimation was similarly applied to dating of gene duplications in Populus and Arabidopsis (Arabidopsis thaliana).
Real-Time RT-PCR Analysis
Relative transcript abundance of all tubulin isoforms in various aspen tissues was analyzed by real-time RT-PCR using the ABsolute QPCR SYBR Green Mix (Abgene) and the Mx3000P real-time PCR system (Stratagene). Gene-specific primers (Supplemental Table S1) flanking 94- to 294-bp amplicons near the 3′-UTRs were designed based on both cloned and JGI-predicted cDNA sequences and, whenever possible, GenBank Populus EST sequences. Each reaction was performed in duplicate with two biological replicates, using cDNA synthesized from 2.5 ng of total RNA. The specificity of amplification was assessed by dissociation curve analysis at the end of each run using the MxPro software (Stratagene), and confirmed by cloning and sequencing of the PCR products, including the PoptrTUA6/8 and PoptrTUB5/6 pairs that could not be distinguished by RT-PCR primers. Relative target transcript levels normalized to the geometric mean of three housekeeping genes (ACTIN, ELONGATION FACTOR1β, and UBIQUITIN) were determined using the ΔCT method (Tsai et al., 2006).
Immunolocalization
Immunolocalization was conducted according to Li et al. (2001) using butyl methyl methacrylate-embedded thin (3-μm) stem sections. Polyclonal anti-TUA antibodies were raised in rabbits using affinity-purified aspen TUA1 recombinant protein (Alpha Diagnostic). The anti-TUB antibodies (ab15568) were obtained from Abcam, and were raised against a synthetic human TUB peptide (416–430) that is highly conserved among PoptrTUBs and corresponds to the C-terminal helix H12 on the outside surface of the MT according to the electron crystallography (Nogales et al., 1998). Anti-TUA antibodies were used at 1/10,000 dilution and anti-TUB serum at 1/2,000 dilution in immunohybridization. Preimmune serum was used as the control. Hybridization signal was colorimetrically detected using nitro blue tetrazolium and 5-bromo-4-chloro-3-indolyl-phosphate. Sections were then mounted in glycerol and images were recorded using a Nikon E-400 microscope equipped with a digital imaging system.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AY229881 and AY229882, EF583813 to EF583816, and EF584828 to EF582849.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Sequence alignment of full-length Populus TUA proteins.
Supplemental Figure S2. Sequence alignment of full-length Populus TUB proteins.
Supplemental Table S1. List of primers used for cloning and real-time RT-PCR analysis.
Supplemental Table S2. List of proteins and their GenBank accession numbers used in sequence alignment and phylogenetic analyses.
Supplementary Material
Acknowledgments
We thank Jingwei Yin for assistance in cloning and sequencing of the TUA6, TUA7, and TUA8 cDNAs; Dr. Vincent Chiang for providing the aspen xylem cDNA library; and Dr. Ramesh Thakur and Dr. David Karnosky for their help with aspen pollen collection.
This work was supported by the U.S. Department of Agriculture National Research Initiative (grant nos. 98–35106–6630 to C.-J.T., 2003–35103–12906 to C.-J.T. and S.A.H., and 2005–35103–15251 to C.-J.T.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Chung-Jui Tsai (chtsai@mtu.edu).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Abe H, Funada R, Ohtani J, Fukazawa K (1995) Changes in the arrangement of microtubules and microfibrils in differentiating conifer tracheids during the expansion of cells. Ann Bot (Lond) 75 305–310 [Google Scholar]
- Abe T, Thitamadee S, Hashimoto T (2004) Microtubule defects and cell morphogenesis in the lefty1lefty2 tubulin mutant of Arabidopsis thaliana. Plant Cell Physiol 45 211–220 [DOI] [PubMed] [Google Scholar]
- Andersson-Gunneras S, Mellerowicz EJ, Love J, Segerman B, Ohmiya Y, Coutinho PM, Nilsson P, Henrissat B, Moritz T, Sundberg B (2006) Biosynthesis of cellulose-enriched tension wood in Populus: global analysis of transcripts and metabolites identifies biochemical and developmental regulators in secondary wall biosynthesis. Plant J 45 144–165 [DOI] [PubMed] [Google Scholar]
- Anthony RG, Hussey PJ (1998) Suppression of endogenous α and β tubulin synthesis in transgenic maize calli overexpressing α and β tubulins. Plant J 16 297–304 [DOI] [PubMed] [Google Scholar]
- Anthony RG, Hussey PJ (1999) Double mutation in Eleusine indica alpha-tubulin increases the resistance of transgenic maize calli to dinitroaniline and phosphorothioamidate herbicides. Plant J 18 669–674 [DOI] [PubMed] [Google Scholar]
- Anthony RG, Reichelt S, Hussey PJ (1999) Dinitroaniline herbicide-resistant transgenic tobacco plants generated by co-overexpression of a mutant alpha-tubulin and a beta-tubulin. Nat Biotechnol 17 712–716 [DOI] [PubMed] [Google Scholar]
- Bhandari S, Fujino T, Thammanagowda S, Zhang D, Xu F, Joshi CP (2006) Coordinate expression of tension stress-responsive and secondary cell wall-associated KORRIGAN endoglucanase and three cellulose synthase genes in aspen trees. Planta 224 828–837 [DOI] [PubMed] [Google Scholar]
- Bondstar JF, Fridovich-Keil JL, Pillus L, Mulligan RC, Solomon F (1986) A chicken-yeast chimeric β-tubulin protein is incorporated into mouse microtubules in vivo. Cell 44 461–468 [DOI] [PubMed] [Google Scholar]
- Brierley HL, Webster P, Long SR (1995) The Pisum sativum TUBa1 gene, a member of a small family of alpha-tubulin sequences. Plant Mol Biol 27 715–727 [DOI] [PubMed] [Google Scholar]
- Burk DH, Ye ZH (2002) Alteration of oriented deposition of cellulose microfibrils by mutation of a katanin-like microtubule-severing protein. Plant Cell 14 2145–2160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai G, Del Casino C, Romagnoli S, Cresti M (2005) Pollen cytoskeleton during germination and tube growth. Curr Sci 89 1853–1860 [Google Scholar]
- Cai G, Romagnoli S, Moscatelli A, Ovidi E, Gambellini G, Tiezzi A, Cresti M (2000) Identification and characterization of a novel microtubule-based motor associated with membranous organelles in tobacco pollen tubes. Plant Cell 12 1719–1736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter JL, Ploense SE, Snustad DP, Silflow CD (1992) Preferential expression of an alpha-tubulin gene of Arabidopsis in pollen. Plant Cell 4 557–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaffey N, Barlow P, Barnett J (1997) Cortical microtubules rearrange during differentiation of vascular cambial derivatives, microfilaments do not. Trees (Berl) 11 333–341 [Google Scholar]
- Chaffey N, Barnett J, Barlow P (1999) A cytoskeletal basis for wood formation in angiosperm trees: the involvement of cortical microtubules. Planta 208 19–30 [DOI] [PubMed] [Google Scholar]
- Chang S, Puryear J, Cairney J (1993) A simple and efficient method for isolating RNA from pine trees. Plant Mol Biol Rep 11 113–116 [Google Scholar]
- Cheng ZG, Snustad DP, Carter JV (2001) Temporal and spatial expression patterns of TUB9, a beta-tubulin gene of Arabidopsis thaliana. Plant Mol Biol 47 389–398 [DOI] [PubMed] [Google Scholar]
- Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD (2003) Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res 31 3497–3500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delye C, Menchari Y, Michel S, Darmency H (2004) Molecular bases for sensitivity to tubulin-binding herbicides in green foxtail. Plant Physiol 136 3920–3932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duckett CM, Lloyd CW (1994) Gibberellic acid-induced microtubule reorientation in dwarf peas is accompanied by rapid modification of an alpha-tubulin isotype. Plant J 5 363–372 [Google Scholar]
- Dutcher SK (2001) The tubulin fraternity: alpha to eta. Curr Opin Cell Biol 13 49–54 [DOI] [PubMed] [Google Scholar]
- Ehlting J, Mattheus N, Aeschliman DS, Li E, Hamberger B, Cullis IF, Zhuang J, Kaneda M, Mansfield SD, Samuels L, et al (2005) Global transcript profiling of primary stems from Arabidopsis thaliana identifies candidate genes for missing links in lignin biosynthesis and transcriptional regulators of fiber differentiation. Plant J 42 618–640 [DOI] [PubMed] [Google Scholar]
- Ersfeld K, Wehland J, Plessmann U, Dodemont H, Gerke V, Weber K (1993) Characterization of the tubulin-tyrosine ligase. J Cell Biol 120 725–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher DD, Cyr RJ (1998) Extending the microtubule/microfibril paradigm. Cellulose synthesis is required for normal cortical microtubule alignment in elongating cells. Plant Physiol 116 1043–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fosket DE, Morejohn LC (1992) Structural and functional organization of tubulin. Annu Rev Plant Physiol Plant Mol Biol 43 201–240 [Google Scholar]
- Fujita M, Saiki H, Harada H (1974) Electron microscopy of microtubules and cellulose microfibrils in secondary wall formation of poplar tension wood fibers. Mokuzai Gakkaishi 20 147–156 [Google Scholar]
- Gaut BS, Morton BR, McCaig BC, Clegg MT (1996) Substitution rate comparisons between grasses and palms: Synonymous rate differences at the nuclear gene ADH parallel rate differences at the plastid gene rbcL. Proc Natl Acad Sci USA 93 10274–10279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge L, Chiang VL (1996) A full length cDNA encoding trans-cinnamate 4-hydroxylase from developing xylem of Populus tremuloides (PGR96–075). Plant Physiol 112 8618883395 [Google Scholar]
- Geisler-Lee J, Geisler M, Coutinho PM, Segerman B, Nishikubo N, Takahashi J, Aspeborg H, Djerbi S, Master E, Andersson-Gunneras S, et al (2006) Poplar carbohydrate-active enzymes. Gene identification and expression analyses. Plant Physiol 140 946–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giddings TH, Staehelin LA (1991) Microtubule-mediated control of microfibril deposition: a re-examination of the hypothesis. In CW Lloyd, ed, The Cytoskeletal Basis of Plant Growth and Form. Academic Press, San Diego, pp 85–100
- Gilmer S, Clay P, MacRae TH, Fowke LC (1999) Tyrosinated, but not detyrosinated, alpha-tubulin is present in root tip cells. Protoplasma 210 92–98 [Google Scholar]
- Gonzalez-Martinez SC, Wheeler NC, Ersoz E, Nelson CD, Neale DB (2007) Association genetics in Pinus taeda L. I. Wood property traits. Genetics 175 399–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertzberg M, Aspeborg H, Schrader J, Andersson A, Erlandsson R, Blomqvist K, Bhalerao R, Uhlen M, Teeri TT, Lundeberg J, et al (2001) A transcriptional roadmap to wood formation. Proc Natl Acad Sci USA 98 14732–14737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himmelspach R, Williamson RE, Wasteneys GO (2003) Cellulose microfibril alignment recovers from DCB-induced disruption despite microtubule disorganization. Plant J 36 565–575 [DOI] [PubMed] [Google Scholar]
- Huang S, Jin L, Du J, Li H, Zhao Q, Ou G, Ao G, Yuan M (2007) SB401, a pollen-specific protein from Solanum berthaultii, binds to and bundles microtubules and F-actin. Plant J 51 406–418 [DOI] [PubMed] [Google Scholar]
- Idriss HT (2000) Man to trypanosome: the tubulin tyrosination/detyrosination cycle revisited. Cell Motil Cytoskeleton 45 173–184 [DOI] [PubMed] [Google Scholar]
- Ko JH, Han KH, Park S, Yang J (2004) Plant body weight-induced secondary growth in Arabidopsis and its transcription phenotype revealed by whole-transcriptome profiling. Plant Physiol 135 1069–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch MA, Haubold B, Mitchell-Olds T (2000) Comparative evolutionary analysis of chalcone synthase and alcohol dehydrogenase loci in Arabidopsis, Arabis, and related genera (Brassicaceae). Mol Biol Evol 17 1483–1498 [DOI] [PubMed] [Google Scholar]
- Kopczak SD, Haas NA, Hussey PJ, Silflow CD, Snustad DP (1992) The small genome of Arabidopsis contains at least 6 expressed alpha-tubulin genes. Plant Cell 4 539–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Nei M (2004) MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform 5 150–163 [DOI] [PubMed] [Google Scholar]
- Ledbetter MC, Porter KR (1963) A “microtubule” in plant cell fine structure. J Cell Biol 19 239–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Cheng XF, Leshkevich J, Umezawa T, Harding SA, Chiang VL (2001) The last step of syringyl monolignol biosynthesis in angiosperms is regulated by a novel gene encoding sinapyl alcohol dehydrogenase. Plant Cell 13 1567–1586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XB, Cai L, Cheng NH, Liu JW (2002) Molecular characterization of the cotton GhTUB1 gene that is preferentially expressed in fiber. Plant Physiol 130 666–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luduena RF (1998) Multiple forms of tubulin: different gene products and covalent modifications. Int Rev Cytol 178 207–275 [DOI] [PubMed] [Google Scholar]
- Lynch M, Conery JS (2000) The evolutionary fate and consequences of duplicate genes. Science 290 1151–1155 [DOI] [PubMed] [Google Scholar]
- MacRae TH (1997) Tubulin post-translational modifications—enzymes and their mechanisms of action. Eur J Biochem 244 265–278 [DOI] [PubMed] [Google Scholar]
- McKean PG, Vaughan S, Gull K (2001) The extended tubulin superfamily. J Cell Sci 114 2723–2733 [DOI] [PubMed] [Google Scholar]
- Montoliu L, Rigau J, Puigdomenech P (1990) A tandem of alpha-tubulin genes preferentially expressed in radicular tissues from Zea mays. Plant Mol Biol 14 1–15 [DOI] [PubMed] [Google Scholar]
- Morejohn LC, Fosket DE (1984) Inhibition of plant microtubule polymerization in vitro by the phosphoric amide herbicide amiprophos-methyl. Science 224 874–876 [DOI] [PubMed] [Google Scholar]
- Morello L, Bardini M, Sala F, Breviario D (2002) A long leader intron of the OsTUB16 rice beta-tubulin gene is required for high-level gene expression and can autonomously promote transcription both in vivo and in vitro. Plant J 29 33–44 [DOI] [PubMed] [Google Scholar]
- Nogales E, Wolf SG, Downing KH (1998) Structure of the αβ tubulin dimer by electron crystallography. Nature 391 199–203 [DOI] [PubMed] [Google Scholar]
- Norberg PH, Meier H (1966) Physical and chemical properties of the gelatinous layer in tension wood fibers of aspen (Populus tremula L.). Holzforschung 20 174–178 [Google Scholar]
- Paredez AR, Somerville CR, Ehrhardt DW (2006) Visualization of cellulose synthase demonstrates functional association with microtubules. Science 312 1491–1495 [DOI] [PubMed] [Google Scholar]
- Pilate G, Dejardin A, Laurans F, Leple JC (2004) Tension wood as a model for functional genomics of wood formation. New Phytol 164 63–72 [DOI] [PubMed] [Google Scholar]
- Prodhan A, Funada R, Ohtani J, Abe H, Fukazawa K (1995) Orientation of microfibrils and microtubules in developing tension-wood fibers of Japanese ash (Fraxinus mandshurica var Japonica). Planta 196 577–585 [Google Scholar]
- Ramakrishna W, Dubcovsky J, Park YJ, Busso C, Emberton J, SanMiguel P, Bennetzen JL (2002) Different types and rates of genome evolution detected by comparative sequence analysis of orthologous segments from four cereal genomes. Genetics 162 1389–1400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers HJ, Greenland AJ, Hussey PJ (1993) Four members of the maize beta-tubulin gene family are expressed in the male gametophyte. Plant J 4 875–882 [DOI] [PubMed] [Google Scholar]
- Romagnoli S, Cai G, Cresti M (2003) In vitro assays demonstrate that pollen tube organelles use kinesin-related motor proteins to move along microtubules. Plant Cell 15 251–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudiger M, Wehland J, Weber K (1994) The carboxy-terminal peptide of detyrosinated alpha tubulin provides a minimal system to study the substrate specificity of tubulin-tyrosine ligase. Eur J Biochem 220 309–320 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW (2001) Molecular Cloning: A Laboratory Manual, Ed 3. Cold Spring Harbor Laboratory Press, Woodbury, NY
- Smertenko A, Blume Y, Viklicky V, Opatrny Z, Draber P (1997) Post-translational modifications and multiple tubulin isoforms in Nicotiana tabacum L. cells. Planta 201 349–358 [DOI] [PubMed] [Google Scholar]
- Snustad DP, Haas NA, Kopczak SD, Silflow CD (1992) The small genome of Arabidopsis contains at least 9 expressed beta-tubulin genes. Plant Cell 4 549–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spokevicius AV, Southerton SG, MacMillan CP, Qiu D, Gan S, Tibbits JFG, Moran GF, Bossinger G (2007) Beta-tubulin affects cellulose microfibril orientation in plant secondary fibre cell walls. Plant J 51 717–726 [DOI] [PubMed] [Google Scholar]
- Stanchi F, Corso V, Scannapieco P, Ievolella C, Negrisolo E, Tiso N, Lanfranchi G, Valle G (2000) TUBA8: a new tissue-specific isoform of alpha-tubulin that is highly conserved in human and mouse. Biochem Biophys Res Commun 270 1111–1118 [DOI] [PubMed] [Google Scholar]
- Sullivan KF (1988) Structure and utilization of tubulin isotypes. Annu Rev Cell Biol 4 687–716 [DOI] [PubMed] [Google Scholar]
- Touchell DH, Wang YS, Harding SA, Tsai CJ (2003) Differential display. In LJ Cseke, PB Kaufman, GK Podila, CJ Tsai, eds, Handbook of Molecular and Cellular Methods in Biology and Medicine, Ed 2. CRC Press, Boca Raton, FL, pp 305–317
- Tsai CJ, Harding SA, Tschaplinski TJ, Lindroth RL, Yuan Y (2006) Genome-wide analysis of the structural genes regulating defense phenylpropanoid metabolism in Populus. New Phytol 172 47–62 [DOI] [PubMed] [Google Scholar]
- Tuskan GA, DiFazio S, Jansson S, Bohlmann J, Grigoriev I, Hellsten U, Putnam N, Ralph S, Rombauts S, Salamov A, et al (2006) The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313 1596–1604 [DOI] [PubMed] [Google Scholar]
- Wasteneys GO (2004) Progress in understanding the role of microtubules in plant cells. Curr Opin Plant Biol 7 651–660 [DOI] [PubMed] [Google Scholar]
- Weinstein B, Solomon F (1990) Phenotypic consequences of tubulin overproduction in Saccharomyces cerevisiae: differences between alpha-tubulin and beta-tubulin. Mol Cell Biol 10 5295–5304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann S, Weber K (2003) Post-translational modifications regulate microtubule function. Nat Rev Mol Cell Biol 4 938–948 [DOI] [PubMed] [Google Scholar]
- Whittaker DJ, Triplett BA (1999) Gene-specific changes in alpha-tubulin transcript accumulation in developing cotton fibers. Plant Physiol 121 181–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa M, Yang GX, Kawaguchi K, Komatsu S (2003) Expression analyses of beta-tubulin isotype genes in rice. Plant Cell Physiol 44 1202–1207 [DOI] [PubMed] [Google Scholar]
- Yoshimura T, Demura T, Igarashi M, Fukuda H (1996) Differential expression of three genes for different beta-tubulin isotypes during the initial culture of Zinnia mesophyll cells that divide and differentiate into tracheary elements. Plant Cell Physiol 37 1167–1176 [DOI] [PubMed] [Google Scholar]
- Zhong R, Burk DH, Morrison WH III, Ye ZH (2002) A kinesin-like protein is essential for oriented deposition of cellulose microfibrils and cell wall strength. Plant Cell 14 3101–3117 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.