Abstract
We have previously shown that the beneficial filamentous fungus Trichoderma virens secretes the highly effective hydrophobin-like elicitor Sm1 that induces systemic disease resistance in the dicot cotton (Gossypium hirsutum). In this study we tested whether colonization of roots by T. virens can induce systemic protection against a foliar pathogen in the monocot maize (Zea mays), and we further demonstrated the importance of Sm1 during maize-fungal interactions using a functional genomics approach. Maize seedlings were inoculated with T. virens Gv29-8 wild type and transformants in which SM1 was disrupted or constitutively overexpressed in a hydroponic system or in soil-grown maize seedlings challenged with the pathogen Colletotrichum graminicola. We show that similar to dicot plants, colonization of maize roots by T. virens induces systemic protection of the leaves inoculated with C. graminicola. This protection was associated with notable induction of jasmonic acid- and green leaf volatile-biosynthetic genes. Neither deletion nor overexpression of SM1 affected normal growth or development of T. virens, conidial germination, production of gliotoxin, hyphal coiling, hydrophobicity, or the ability to colonize maize roots. Plant bioassays showed that maize grown with SM1-deletion strains exhibited the same levels of systemic protection as non-Trichoderma-treated plants. Moreover, deletion and overexpression of SM1 resulted in significantly reduced and enhanced levels of disease protection, respectively, compared to the wild type. These data together indicate that T. virens is able to effectively activate systemic disease protection in maize and that the functional Sm1 elicitor is required for this activity.
Plants in their natural settings are surrounded by a range of beneficial or deleterious microorganisms. By evolving mechanisms that enable recognition of the invader followed by production of an arsenal of antimicrobial and/or antiherbivory compounds, plants have the ability to localize and reduce the impact of the pathogen invasion (Paul et al., 2000; Veronese et al., 2003). One of the most effective resistance mechanisms, an induced defense response, is activated only upon pathogen invasion and involves multiple signal transduction pathways. This strategy appears more cost effective than constitutive defenses as these pathways are only activated upon invasion (Heil and Bostock, 2002). Deployment of the plant defenses occurs both locally, at the site of infection, and/or systemically, e.g. in tissues distant from the point of pathogen entry (Gozzo, 2003). A number of plant species have been shown to develop systemic acquired resistance (SAR) in response to local infection with necrotizing pathogens conferring a broad-range resistance (Durrant and Dong, 2004; Glazebrook, 2005). SAR is characterized by an early increase in endogenous salicylic acid (SA) and accumulation of pathogenesis-related (PR) proteins (Van Loon and Van Strien, 1999; Durrant and Dong, 2004). Interestingly, induced systemic responses are not just initiated by pathogens, but also may result from interactions with avirulent microbes. The colonization of the rhizosphere by certain strains of plant growth-promoting rhizobacteria (PGPR) results in a state of heightened resistance to subsequent pathogen attack, a phenomenon generally known as induced systemic resistance (ISR; van Loon et al., 1998; Pieterse et al., 2003). Rhizobacteria-mediated ISR can occur in many plant species and was also demonstrated to be effective against a broad range of pathogens (van Loon et al., 1998). Unlike SA-dependent SAR, extensive work with Arabidopsis (Arabidopsis thaliana) has demonstrated that ISR depends primarily on signal transduction pathways involving jasmonic acid (JA) and ethylene (ET; Ton et al., 2002). Recent findings indicate that there is cross talk and a certain degree of overlap between SA and JA signaling pathways (Schenk et al., 2000; Dong, 2001; Glazebrook et al., 2003).
In addition to plant growth-promoting rhizobacteria, there is another group of root-colonizing beneficial microorganisms that have been found to induce plant resistance to pathogens. They represent the anamorphic stages of several fungi, including Trichoderma spp., Fusarium spp., binucleate Rhizoctonia, and Pythium oligandrum, and are commonly found in most soils throughout the world (Hwang and Benson, 2003; Harman et al., 2004a; Le Floch et al., 2005). Members of the genus Trichoderma have long been recognized as agents for the biocontrol of plant diseases. It is now widely accepted that the biocontrol properties of Trichoderma spp. are also based on their ability to induce both local and systemic resistance responses. To date, however, the induction mechanisms as well as the fungal elicitors and the plant signals involved are largely undefined. Previous studies have reported that the root colonization by Trichoderma spp. resulted in the accumulation of antimicrobial compounds both locally in the roots (Howell et al., 2000) and systemically in the leaves (Yedidia et al., 2003). More recently, a JA/ET signaling pathway and a mitogen-activated protein kinase signaling pathway of both the plant and the fungus were identified to be important for the Trichoderma spp.-mediated ISR in cucumber (Cucumis sativus) plants (Shoresh et al., 2005, 2006; Viterbo et al., 2005). However, fungal elicitors and/or the plant signaling compounds related to these pathways still await discovery.
The highly coordinated molecular dialogue that occurs between plants and microbes during the early stages of their association, in which signaling molecules play an essential role, determines the final outcome of the relationship, which ranges from parasitism to mutualism (Bais et al., 2004; Pozo et al., 2005). A large array of microbial elicitors that initiate plant defense responses has been characterized (for review, see Nimchuk et al., 2003). Plant cells exposed to elicitors (i.e. crude fungal cell wall fragments or defined molecules such as purified proteins and avirulence gene products) respond with a battery of cellular changes, including rapid ion fluxes and the generation of reactive oxygen species, accumulation of phytoalexins, and synthesis of PR proteins (Nicholson and Hammerschmidt, 1992; Van Loon and Van Strien, 1999; Mittler et al., 2004). However, a clear understanding of the Trichoderma-plant recognition and communication process is lacking. Only proteins with enzymatic activity (i.e. cellulase and xylanase) have been described as proteinaceous elicitors in Trichoderma spp. (Bailey et al., 1992; Calderon et al., 1993). It should be noted that in case of xylanase, it was demonstrated that its enzymatic activity is unrelated to the elicitation processes (Furman-Matarasso et al., 1999; Rotblat et al., 2002). Although evidence exists indicating that other metabolites capable of elicitation of plant defense are produced by Trichoderma (Hanson and Howell, 2004; Harman et al., 2004a), these proposed elicitors have not been characterized.
To obtain new insights into the events underlining the processes of plant-Trichoderma interactions, we previously identified and characterized an elicitor produced by Trichoderma virens named Sm1, a novel proteinaceous nonenzymatic elicitor from this group of rhizosphere-competent fungi (Djonovic et al., 2006a). Sm1 is induced and secreted by the fungus at the early stages of plant-Trichoderma interaction, suggesting a signaling role for this protein. Indeed, the purified Sm1 efficiently elicited plant defense responses and systemic resistance against a foliar pathogen of cotton (Gossypium hirsutum; Djonovic et al., 2006a). The protective activity of Sm1 was associated with the accumulation of reactive oxygen species and phenolic compounds, and increased levels of transcription of the defense genes regulated by SA and JA/ET as well as genes involved in the biosynthesis of sesquiterpenoid phytoalexins (Djonovic et al., 2006a).
Most of the molecular mechanisms that underlie the Trichoderma-mediated systemic induced resistance have been studied in dicot plants (Yedidia et al., 2003; Shoresh et al., 2005; Viterbo et al., 2005; Djonovic et al., 2006a). In contrast, the current knowledge of SAR/ISR mechanisms in monocots is limited. The only available data suggesting that monocots may undergo induced resistance responses similar to those found in dicots have been generated from the exogenous application of chemical inducers of SAR (Kogel et al., 1994; Schweizer et al., 1999). However, conclusive evidence of the biological relevance of this chemically induced SAR is lacking. Moreover, unlike multiple reports on dicot species, the monocot “SAR genes” that are inducible by pathogens were not inducible by chemicals (Schaffrath et al., 1997; Schweizer et al., 1999). It is worth noting, however, that despite these differences, many lines of evidence point to a similarity in various aspects of induced resistance between monocots and dicots (Morris et al., 1998; Dong, 2004; Chern et al., 2005).
In this study we aimed (1) to determine whether colonization of roots by T. virens can induce systemic protection against a foliar pathogen in the monocot maize (Zea mays); (2) to examine the requirement of a functional Sm1 for induction of systemic resistance in maize; (3) to elucidate the potential involvement of signaling pathways mediated by SA, JA, and other oxygenated fatty acids in the induced resistance of maize triggered by T. virens; and (4) to assess a number of phenotypic traits to address a potential role of Sm1 in the physiology of T. virens. This study provides compelling evidence that, similar to dicot plants, colonization of maize roots by T. virens induces ISR-like systemic protection of the leaves inoculated with Colletotrichum graminicola. Furthermore, our results demonstrate that Sm1 is required for activation of ISR in maize, since plants grown with SM1-deletion strains exhibited the same levels of systemic protection as the control (non-Trichoderma-treated) plants and, conversely, plants grown with SM1-overexpression strains displayed increased protection compared to the wild type.
RESULTS
Identification of SM1-Deletion and -Overexpression Transformants
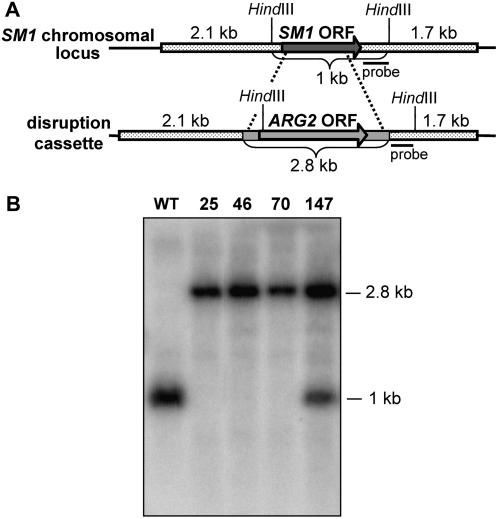
Previously, we showed that a T. virens hydrophobin-like elicitor, Sm1, induced plant defense responses and provided high levels of systemic resistance against the foliar pathogen Colletotrichum sp. in cotton (Djonovic et al., 2006a). To confirm the biological relevance of Sm1 in the T. virens Gv29-8 (wild type) systemic induced resistance in maize, we generated SM1-deletion (KO) and -overexpression (OE) strains. The vector for gene replacement, pSZD25, was constructed to replace the entire SM1 open reading frame (ORF) with a selectable marker (Fig. 1A). A total of 147 stable transformants were tested for a gene disruption by PCR (data not shown). Nine PCR-selected candidates were further analyzed by Southern hybridization to verify the gene disruption. After digestion of the genomic DNA with HindIII and hybridization with a 460-bp BamHI/HindIII fragment immediately downstream of SM1, a positive disruption event yielded a 2.8-kb band versus a 1-kb band in the wild type (Fig. 1A). Three strains, KO25, KO46, and KO70, were clearly disrupted in the gene, and strain 147 contained both 1-kb and 2.8-kb bands, indicating ectopic integration of pSZD25 (Fig. 1B). After probing with a 710-bp XhoI fragment from the ARG2 gene, a single integration event was detected in disruption transformants (data not shown).
Figure 1.
Southern analysis and confirmation of SM1 disruptants. A, Scheme of the gene-deletion strategy. The expected sizes in native (1 kb) and deletion (2.8 kb) events are indicated. The dimensions are not drawn to scale. B, Southern analysis of T. virens wild-type (WT) strain and SM1-deletion transformants (KO25, KO46, and KO70). Autoradiograph of DNA gel blot hybridized with 32P-dCTP-labeled probe indicated in A. Fifteen micrograms of genomic DNA was digested with HindIII and loaded per lane. Numbers on the right indicate expected size in native and deletion events. Strain 147 contains the native and overexpression bands indicating ectopic integration.
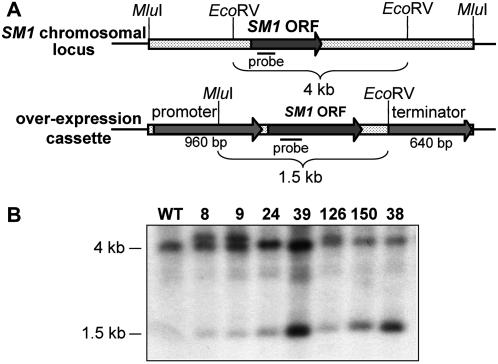
The overexpression vector pSZD26 was constructed for constitutive overproduction of the SM1 gene (Fig. 2A). From 14 randomly selected putative overexpression transformants, 10 demonstrated the presence of the overexpression cassette based on PCR analysis (data not shown). These 10 candidates were further verified by Southern-blotting analysis. As Figure 2 demonstrates, the SM1 probe hybridized to the native gene, yielding a 4-kb and a 1.5-kb band in overexpression strains, as expected (Fig. 2B). The intensity of hybridization of the 1.5-kb band in some of the transformants, such as OE38 and OE39, suggests integration of multiple copies of the construct at one site, resulting in tandem repeats.
Figure 2.
Confirmation of SM1-overexpression transformants. A, Gene-overexpression strategy. The expected sizes in native (4 kb) and overexpression (1.5 kb) events are indicated. The dimensions are not drawn to scale. B, Southern analysis of T. virens wild-type (WT) strain and SM1-overexpression transformants (OE8, OE9, OE24, OE39, OE126, OE150, and OE38). Fifteen micrograms of genomic DNA was double digested with MluI/EcoRV, blotted on a membrane, and probed with the 32P-dCTP-labeled 264-bp PCR product amplified from SM1 ORF. Numbers on the left indicate expected size in overexpression and wild-type sequences.
Expression of SM1 in Transformants
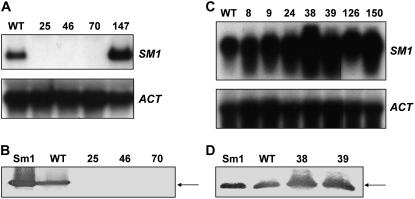
Gene expression analysis was performed to test whether disruption or overexpression of the SM1 gene resulted in the expected alteration of transcript and protein accumulation. Northern-blot experiments showed no transcripts of SM1 in any of the deletion transformants (KO25, KO46, or KO70; Fig. 3A). These results were further verified by western-blot analysis (Fig. 3B), confirming the successful replacement of the gene. Additionally, there was no obvious difference in the extracellular protein profile of the wild-type and deletion strains except for the absence of 12.6-kD Sm1 band in deletion strains (data not shown). Examination of the seven overexpression strains (OE8, OE9, OE24, OE38, OE39, OE126, and OE150) revealed higher SM1 mRNA levels than for the wild type after 3 d of growth in a complete medium (GYEC; Fig. 3C). Polypeptide levels for the overexpression strains (OE38 and OE39) with the highest SM1 mRNA levels were compared to the wild type by western-blot analysis. Both overexpression stains produced greater amounts of Sm1 than the wild type (Fig. 3D).
Figure 3.
SM1 expression in transformants. A and C, Northern analysis of transformants. Total RNA was extracted from wild type (WT), deletion strains (SKO25, SKO46, SKO70), and strain with ectopic integration of the construct (147) cultured in VMS media for 5 d (A), and overexpression strains (OE8, OE9, OE24, OE38, OE39, OE126, and OE150) cultured in GYEC medium for 3 d (C). Fifteen micrograms in A and 10 μg in C of total RNA were separated on formaldehyde-agarose gel, transferred to a Hydrobond-N+ nylon membrane, and hybridized with the 264-bp PCR product of the SM1 gene (A and C, top). The same membrane was hybridized with a 550-bp actin fragment PCR amplified from T. virens wild-type cDNA as a control for even loading (A and C, bottom). B and D, Western analysis of transformants. Immunoblot of the protein profile of 5-d VMS CFs from T. virens wild-type and deletion strains (B) and 3-d CFs of overexpression and wild-type strains cultured in GYEC medium (D). Pure Sm1 was included as positive control (Sm1). Six micrograms of total protein of each strain was loaded per lane. The 12.6-kD Sm1 is indicated by arrows.
Phenotypic Analysis of Transformants
A number of phenotypic assays were conducted with the gene-disruption, gene-overexpression, and wild-type strains to assess if the morphology and physiology of strains were affected by genetic manipulations of SM1.
There were no evident changes in the cultures of the deletion or overexpression transformants when compared to the wild type with respect to colony appearance and pigmentation during sporulation. We further assessed the possible involvement of SM1 in fungal growth under different nutritional conditions. Three disruptants, KO25, KO46, and KO70, and two overexpression strains, OE38 and OE39, were selected for growth analysis. Growth area was compared after 1 and 2 d growth on minimal (Vogel's minimal medium [Vogel, 1956] supplemented with 1.5% Suc [VMS], water agar [WA]) or complex (potato dextrose agar [PDA]) medium. An ANOVA (P < 0.05) indicated there was no significant differences in growth area among the strains on any of the media tested (Supplemental Table S1). Additionally, no differences among the strains were found in conidial germination. The percentage of germination ranged from 92.3% to 93.7% and was not significantly different. The strains were also assessed for their ability to coil around their target fungal hosts as a component of the mycoparasitic process (Benitez et al., 2004). The ability to recognize and attach and coil around the fungal host Rhizopus oryzae was not affected by either SM1 deletion or overexpression (Supplemental Fig. S1). Zones of inhibition on antibacterial indicator plates resulting from the presence of gliotoxin in culture filtrates (CFs) were similar among all strains tested.
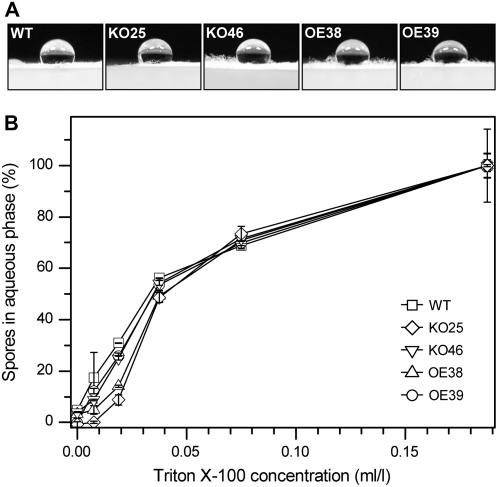
Since Sm1 is a hydrophobin-like protein, we further assessed the hydrophobicity of transformants by performing two different assays. In the first experiment, the hydrophobicity of the mycelia was visualized by placing water droplets on the T. virens strains growing on a solid agar medium (Fig. 4A). The greater the contact angle of a droplet, the greater the hydrophobicity and vice versa (van der Mei et al., 1991). Figure 4A shows that the hydrophobicity of the wild-type strain and the SM1-deletion or -overexpression strains did not visibly differ. The second assay involved separation of conidia into aqueous and organic phases to assess hydrophobicity of conidia (Whiteford and Spanu, 2001). Titrating conidia from this interface with a detergent (e.g. Triton X-100) can release conidia and may reveal quantifiable differences in conidium hydrophobicity between strains (Fig. 4B). From the Figure 4B we can see that the amount of detergent needed to release the conidia of wild-type, SM1-deletion, and SM1-overexpression strains into the aqueous phase was very similar. The absence of any visible alteration of the fungal growth provided confidence that subsequent results from the biocontrol assays were a function of genetic manipulation in SM1 rather than the transformation procedure.
Figure 4.
Hydrophobicity of wild-type, SM1-deletion, and SM1-overexpression strains. A, Hydrophobicity of wild-type (WT), SM1-deletion (KO25 and KO46), and SM1-overexpression strains (OE38 and OE39). Water droplet (20 μL) was placed on the surface of the colony of each strain after 2 d growth on PDA (nonsporulating mycelium) and photographed. B, Hydrophobicity of conidia using MATH assay (Whiteford and Spanu, 2001). The graph shows the amount of conidia in the aqueous phase in the presence of increasing concentrations of the surfactant Triton X-100 (each point is the mean of two replicates).
Genetic Manipulation of SM1 Results in Altered Local and Systemic Expression of a Defense-Related PAL Gene in Hydroponically Grown Maize
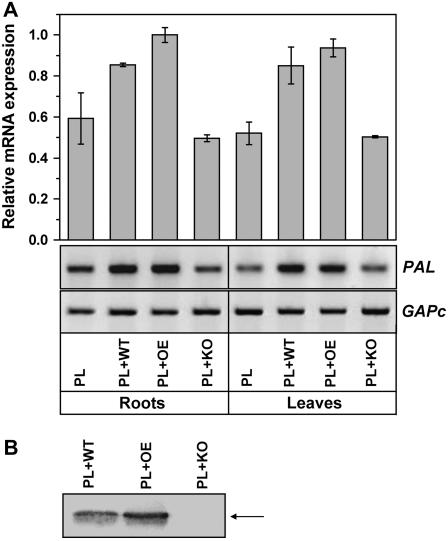
We have previously shown that colonization of cotton roots by T. virens resulted in an increased expression of PR genes (Djonovic et al., 2006a). To investigate plant defense responses to treatment of roots with T. virens in a monocot species, maize (inbred line B73) was selected as a model plant. An aseptic hydroponic growth system was used (Djonovic et al., 2006a) to spatially separate the root-inducing agent from the distal cotyledon tissue. To verify that the fungal strains did not colonize the entire seedling, maize aboveground organs were plated onto a Trichoderma selective medium (GVSM; Park et al., 1992). The presence of T. virens was not detected in any of the maize aboveground organs. Expression of maize defense genes was tested in root and leaf tissues at 48 h after inoculation of the root system with the wild type or transformants. Figure 5A illustrates that defense-related PAL (Phe ammonia-lyase) gene was up-regulated locally, in roots, and systemically, in leaves, when maize seedlings were grown with the wild-type and overexpression strains. In contrast, no significant changes in the levels of expression of PAL were detected in either roots or leaves of seedlings cocultured with the deletion strains (Fig. 5A). These results indicated that the functional Sm1 elicitor is required for induction of the plant PAL gene.
Figure 5.
sqRT-PCR analysis of PAL expression in maize hydroponically grown with T. virens strains and Sm1 expression. Shown is PAL expression in maize root and leaf tissue 48 h after root inoculation with T. virens wild type (PL + WT), SM1-overexpression strain OE38 (PL + OE), or SM1-deletion strain KO25 (PL + KO). Plants grown without Trichoderma were included as control (PL). Maize GAPc was used to as control for equal amounts of cDNA. Intensities of bands were quantified using Scion Image software and the data expressed as arbitrary units of PAL expression normalized to GAPc. The values are shown relative to the highest value (bars) and are representative of two independent experiments with se bars. B, Immunoblot analysis of Sm1 secreted into the growth medium in the hydroponically cocultured Trichoderma-maize seedlings. Equal volumes of concentrated samples equivalent to 300 mL growth medium from the hydroponic system (PL + WT, PL + OE, PL + KO) were loaded on a 15% SDS-PAGE and electroblotted to a nitrocellulose membrane. The 12.6-kD Sm1 is indicated by an arrow.
Detection of Sm1 during Fungal-Maize Interaction in the Hydroponic System
As we have previously shown that Sm1 was abundantly secreted outside the cell in the presence of cotton seedlings (Djonovic et al., 2006a), we performed western analysis to examine the expression of Sm1 in wild-type and overexpression strains in the hydroponically cocultured Trichoderma-maize seedlings (Fig. 5B). The analysis of the secreted proteins in the plant growth medium showed that Sm1 was more abundant in the filtrate from the plant-overexpression strain coculture than in the filtrate from the plant-wild type coculture (Fig. 5B). As expected, no Sm1 was detected in the plant-deletion strain coculture (Fig. 5B).
Sm1 Is Required for ISR in Maize against C. graminicola
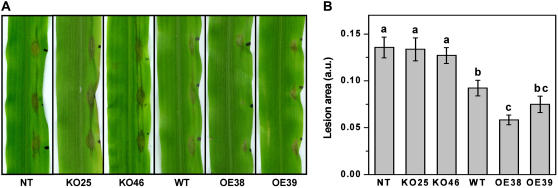
To assess the in vivo biological relevance of T. virens to induce disease resistance in maize through Sm1 expression, wild-type strain, two deletion strains, and two overexpression strains were compared in their ability to systemically protect 2-week-old maize seedlings challenged with the foliar pathogen C. graminicola. Maize seedlings from T. virens-treated seed were inoculated with a spore suspension of the pathogen with disease assessment conducted 4 d following inoculation. In contrast to the typical large lesions that appeared earlier and had began to coalesce in the plants not treated with T. virens or plants treated with SM1-deletion strains, the lesions were significantly smaller in plants treated with either wild-type strain or SM1-overexpression strains. Disease protection due to treatment with overexpression strains was even more evident than with the wild type, as the lesions were much smaller, and development of some lesions was delayed or arrested (Fig. 6A). The mean lesion area for the plants treated with the disruptant strains was significantly greater than wild-type or overexpression strains, but was the same as the nontreated control. Both overexpression strains increased disease protection as compared to the wild-type strain with OE38 significantly reducing lesion area (P < 0.05; Fig. 6B).
Figure 6.
Effect of T. virens wild-type, SM1-deletion, and SM1-overexpression strains on ISR in maize seedlings to C. graminicola. A, Lesion development in leaves of T. virens-induced maize plants 4 d postchallenge with the pathogen C. graminicola. From left to right: control seedlings non-Trichoderma inoculated (NT), and seedlings inoculated with SM1-deletion (KO25 and KO46), T. virens wild-type (WT), and SM1-overexpression (OE38 and OE39) strains. B, The graph illustrates the levels of systemic disease protection observed in each treatment. The mean lesion area (given as the arbitrary units) was evaluated 4 d postinoculation as described in “Materials and Methods.” Each bar represents mean lesion area of six replicates with four plants each from three independent experiments with se bars. Columns with letter in common did not differ significantly according to Fisher's PLSD test at a significance level of 5%.
Colonization of Roots by T. virens Strains in Hydroponics and in Soil Is Not Affected by SM1 Deletion or Overexpression
As Sm1 is a hydrophobin-like protein and hydrophobins are known to be involved in several plant-fungal symbiotic interactions (Tagu et al., 2002; Duplessis et al., 2005; Viterbo and Chet, 2006), we hypothesized that the inability of T. virens to produce Sm1 may have an affect on the ability of these strains to colonize plant roots. However, our data show that neither deletion nor overexpression of SM1 affected the ability of the wild-type strain to colonize seedlings grown hydroponically or in soil (Table I).
Table I.
Colonization by wild-type (WT), SM1-deletion (KO), or SM1-overexpression (OE) strains of maize seedlings grown in hydroponic system or in soil
| Strain | Hydroponicsa | Soilb |
|---|---|---|
| WT | 94.40 ± 1.88 | 1.92 ± 0.21 |
| KO25 | 97.25 ± 0.64 | 2.57 ± 0.21 |
| KO46 | 96.15 ± 1.29 | 1.96 ± 0.15 |
| OE38 | 95.02 ± 0.91 | 1.98 ± 0.38 |
| OE39 | 94.83 ± 1.46 | 2.12 ± 0.32 |
The percentage of the total length of excised root fragments from the hydroponic system colonized by the strains of T. virens given as a mean of six replicates with the se.
Total number of colonies of the strains per centimeter of root harvested from the maize seedling grown in soil given as a mean of five replicates with the se. These experiments were replicated at least once with similar results. Based on ANOVA (P < 0.05), there was no significant differences in root colonization in any of the growth systems.
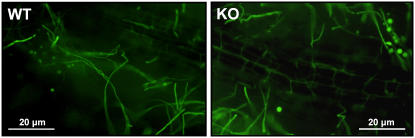
To further examine the colonization of maize seedlings roots by the wild-type and SM1-deletion strains, we generated T. virens wild-type and deletion strains expressing GFP. Southern analysis of genomic DNAs of transformants confirmed integration of the GFP and HPH (hygromycin B phosphotransferase gene) vectors, pTEFEGFP and pCSN44, respectively (data not shown). The growth rate and colony morphology of selected cotransformants were similar to the original strains as evaluated by growth comparison on various nutritional media (PDA, VMS, and WA plates). Six-day-old maize seedlings grown in the hydroponic system were examined for the pattern of colonization by these strains. The extent of colonization (fungal hyphae along the main root axis as well as encircling the roots) as well terminal and intercalary formation of chlamydospores appeared similar for both strains (Fig. 7).
Figure 7.
Fluorescent micrographs of 6-d-old maize seedlings grown in the hydroponic system and colonized with T. virens wild-type or deletion strain (KO25) expressing GFP. Microscopy was performed using Olympus BX-51 fluorescent microscope with excitation from 470 to 490 nm, emission from 510 to 550 nm, and 200× magnification.
Expression of Maize Defense Genes during T. virens-Induced Resistance to C. graminicola
To elucidate the potential involvement of signaling pathways mediated by SA, JA, and other octadecanoids in the induced resistance of maize triggered by T. virens, we examined the expression of nine well-characterized maize defense genes. The housekeeping gene, the cytosolic form of glycerol phosphate dehydrogenase (GAPc; Farag et al., 2005), was used to ensure equal amount of template cDNA. Because many of the genes tested belong to large gene families and share high sequence identity throughout their entire cDNA, gene-specific primers were generated and semiquantitative reverse transcription (sqRT)-PCR analyses were performed. A range of PCR reactions was conducted to determine the optimal number of PCR cycles for linear amplification of the genes. Fourteen days postinoculation with T. virens wild-type or transformant strains, maize seedlings were inoculated with C. graminicola. Systemic induced resistance was evaluated before pathogen inoculation (T = 0 or equivalent of 14 d growth) or 1 d after pathogen inoculation (T = 1). Total RNA was extracted from the leaf tissue at these time points and sqRT-PCR performed. As the expression of all of the genes studied after pathogen attack, in general, did not differ significantly among Trichoderma-treated or nontreated plants but was inducible by C. graminicola infection (M.V. Kolomiets, unpublished data), we have shown the expression of the maize defense genes activated by the presence of the T. virens strains only (T = 0; Fig. 8).
Figure 8.
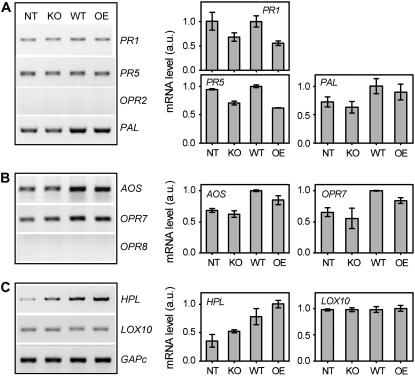
Expression (sqRT-PCR) of defense-related genes in leaves of T. virens wild type-, deletion strain-, and overexpression strain-induced maize plants challenged with C. graminicola. A, SA-responsive genes: PR1 and PR5, OPR2, and PAL. B, JA-biosynthetic or -inductive genes: AOS, OPR7, and OPR8. C, GLV-biosynthetic or -inductive genes: HPL and LOX10. Expression of GAPc was used as a quantitative standard. The negative control for sqRT-PCR was DNase-treated and cleaned RNA reactions without reverse transcription (not shown). Treatments are: maize seedlings not inoculated with T. virens (NT), or preinoculated with KO25 (KO), Gv29.8 (WT), or OE38 (OE) for 14 d (T = 0, before pathogen challenge). Intensities of bands were quantified using Scion Image software (right panels) and the data expressed as arbitrary units of expression of the indicated defense genes normalized to GAPc. The values shown are the average of two independent experiments with se bars (note: the images of gene expression on the left are from those of the second experiment).
The maize genes of the SA-responsive pathway examined were PR1 and PR5 (Morris et al., 1998), oxo-phytodienoate reductase OPR2 (Zhang et al., 2005), and PAL (Farag et al., 2005). PR1 and PR5 in the treated plants were expressed at similar levels as the nontreated (control) plants. OPR2 transcripts were not detected at this time point. For PAL, there was a notable up-regulation by wild-type and overexpression strains, whereas expression was similar in nontreated and deletion strain-treated plants (Fig. 8A).
Since lipoxygenase (LOX) products such as jasmonates and other oxygenated fatty acids called oxylipins were implicated as signals in ISR (Conrath et al., 2006), the expression of the LOX pathway (Fig. 8B) was examined. Currently, the best studied branches of this pathway are those initiated by the oxygenation of linolenic acid by 13-LOXs: the allene oxide synthase (AOS) branch and the hydroperoxide lyase (HPL) branch (Feussner and Wasternack, 2002). The AOS branch involves a series of enzymes including allene oxide cyclase and 12-oxo-phytodienoic acid reductase (OPR) and produces the diverse group of jasmonates (Feussner and Wasternack, 2002). The HPL branch final products include a number of volatile C-6 aldehydes and alcohols, or so-called green leaf volatiles (GLVs) that function as signaling molecules in defense responses to pathogens and pests (Matsui, 2006). AOS and OPR7 and/or OPR8 are implicated in JA biosynthesis (Zhang et al., 2005; Mei et al., 2006). Similar to the PAL gene, the expression of AOS and OPR7 was notably up-regulated by wild-type and overexpression strains, whereas a basal level of expression was detected in either nontreated or deletion strain-treated plants. Interestingly, OPR8 was not expressed at any of the stages or treatments tested, suggesting a differential regulation of this other putative JA-producing OPR isoform (Fig. 8B).
LOX10 and HPL are most likely GLV-producing enzymes in maize (Nemchenko et al., 2006). Moreover, LOX10 is the closest maize homolog of a bean (Phaseolus vulgaris ‘Prelude’) LOX gene associated with ISR by a nonpathogenic Pseudomonas strain (Ongena et al., 2004). In our study, LOX10 was expressed at very low levels throughout the time course, and did not differ among nontreated and Trichoderma-treated plants (Fig. 8C). In contrast, the expression of HPL was significantly up-regulated by wild-type and overexpression strains, and expressed at similar levels in deletion strain-treated and nontreated plants. Interestingly, HPL transcript levels were notably higher in overexpression strain treatment compared to the wild type (Fig. 8C).
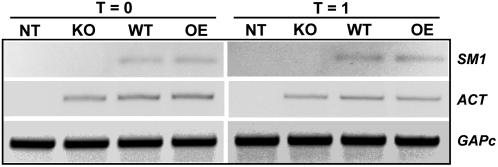
SM1 Is Expressed in Planta Before and After Pathogen Attack
It has been shown recently that several proteins that are involved in Trichoderma-plant early interaction are up-regulated during the interaction with plants but also expressed in planta (Viterbo et al., 2004; Viterbo and Chet, 2006). We have also shown that SM1 was up-regulated in the presence of the cotton plants in the early interaction (Djonovic et al., 2006a). In this study, after observing the protection of maize seedlings pretreated with T. virens wild-type and overexpression strains against a foliar pathogen, we wanted to test if SM1 was indeed expressed in planta after the root colonization by T. virens strains. Figure 9 shows that SM1 was expressed both after 14 d of growth with Trichoderma (Fig. 9, left) as well as after pathogen attack (Fig. 9, right). As expected, no transcripts of SM1 were detected in seedlings grown with SM1-deletion strain or non-Trichoderma-treated plants (Fig. 9). However, the technique used for this experiment was not quantitative (RT-PCR performed with 40 cycles as the level of expression was low), but was employed to confirm that SM1 was expressed in planta in both wild-type and overexpression strains during T. virens colonization of healthy maize seedlings as well as during the infection process.
Figure 9.
Detection of SM1 expression in maize roots (RT-PCR). A, Total RNA was extracted from tergitol-metaphosphate-washed roots of maize seedlings not inoculated with T. virens (NT), or preinoculated with KO25, wild type, or OE38 for 14 d (T = 0, before pathogen challenge) or 1 d after C. graminicola challenge (T = 1). Gene-specific primers for T. virens SM1 and ACT are indicated on the right. GAPc gene was amplified as a reference for equal amounts of maize cDNA.
DISCUSSION
T. virens Induces Systemic Disease Resistance in Maize
Resistance responses triggered by defense-related signal compounds such as SA or its chemical analogs have been described in maize (Morris et al., 1998). There is no evidence, however, that these chemically induced responses result in systemic protection of plants against pathogens. Additionally, there are only a few reports demonstrating that the colonization of roots of any monocot species by beneficial microorganisms induces systemic disease resistance to leaf pathogens (Harman et al., 2004b; Waller et al., 2005). This study provides evidence that T. virens induces systemic resistance to the fungal foliar pathogen C. graminicola in maize as demonstrated by significant reduction in disease symptoms in plants grown with T. virens Gv29-8 compared to nontreated plants. A previous report indicated that T. virens strains G6, G11, and G6-5 were able to induce local but not systemic resistance in cotton (Howell et al., 2000). We have now demonstrated that T. virens Gv29-8 as well as an exogenous application of Sm1 elicitor produced by Gv29-8 (Djonovic et al., 2006a) are able to effectively activate ISR (Fig. 6).
The important component of local and systemic resistance induced by Trichoderma asperellum in cucumber plants (Yedidia et al., 2000, 2003) was shown to be mediated by the ability of Trichoderma hyphae to penetrate several epidermal layers and colonize the intercellular spaces (Yedidia et al., 1999). This colonization was accompanied by increased levels of plant defense transcripts and phytoalexin production. We have previously demonstrated that T. virens Gv29-8 is rhizosphere-competent on cotton (Park et al., 1992) and that colonization of cotton roots by T. virens Gv29-8 resulted in increased expression of PR genes (Djonovic et al., 2006a). Here, we report that T. virens Gv29-8 is able to successfully colonize maize root systems produced in either an aseptic hydroponic system or soil, resulting in a systemic disease resistance accompanied with increased levels of expression of defense-related genes.
Sm1 Is Required for the Ability of T. virens to Induce Systemic Resistance in Maize
As we have shown that T. virens induces systemic resistance in maize, we further tested the hypothesis that deletion of SM1 in T. virens results in attenuated defense responses reducing the levels of protection against pathogen attack. Results from biological assays performed in this study support this hypothesis. We show that SM1-disruptant strains appear not to trigger ISR as plants grown with deletion strains exhibited similar levels of symptoms as control plants (non-Trichoderma-treated plants). Moreover, this effect was further supported by our results showing that SM1-overexpression strains induced even higher levels of protection than wild-type strain-treated plants (Fig. 6). The enhanced levels of protection with overexpression strain-treated plants may have resulted from the higher levels of production of Sm1 in overexpression strains compared to the wild type. This observation is in agreement with our previous finding that Sm1 secreted by the wild type was up-regulated in the presence of the plants compared to the growth in culture medium (Murashige and Skoog + 0.05% Suc) without the plants (Djonovic et al., 2006a). The same observation was made in this study, when maize seedlings were hydroponically cocultured with T. virens in Murashige and Skoog medium (data not shown).
All together, these data indicate that Sm1 is a key player for T. virens-induced systemic disease resistance in maize. To date, most studies report the effect of exogenous application of purified microbial elicitor on plant resistance. This study provides genetic evidence that an elicitor of defense response is required for ISR.
T. virens ISR in Maize Is Modulated through JA- and GLV- Rather Than SA-Regulated Defense Pathways
Since we demonstrated that root colonization of maize plants by T. virens Gv29-8 triggered systemic disease resistance, we sought to determine which defense-related pathways were associated with the observed protection. Considerable progress has been made recently to elucidate the molecular mechanisms involved in Trichoderma-induced systemic resistance; however, most of this work has been performed with dicot plants (Yedidia et al., 2003; Shoresh et al., 2005; Viterbo et al., 2005; Djonovic et al., 2006a). The signal transduction pathways and the defense genes involved in the resistance mechanisms in dicot plants may differ from those in monocot plants. For instance, in dicot plants, JA was suggested to mediate induced resistance through a distinct, SA-independent signaling pathway, and the most common markers of SA-mediated signal transduction pathways are PR genes (Pieterse and van Loon, 1999). In monocots, accumulating evidence suggests that both signal pathways contribute simultaneously to resistance mechanisms. In maize, PR1 and PR5 genes can be induced by pathogen infection and functional mimics of SA (Morris et al., 1998), whereas in rice (Oryza sativa) exogenous application of JA induces the expression of a number of PR genes (Agrawal et al., 2000; Rakwal and Komatsu, 2000). Moreover, a recent report provided evidence of the importance of JA in mediating rice PR gene expression and disease resistance (Mei et al., 2006). In our study, PR1 and PR5 were expressed at similar levels in the control (noninoculated) and T. virens wild-type strain-inoculated plants, suggesting that these two genes are not involved in T. virens-mediated systemic resistance in maize. This is in agreement with a recent report (Waller et al., 2005), in which systemic disease protection of barley (Hordeum vulgare) infected with the endophytic fungus Piriformospora indica was not accompanied by increased levels of PR5. The potential involvement of the SA-mediated pathway by T. virens was further examined by evaluating the expression of the OPR2 gene, previously shown to be inducible by SA and pathogens but not by JA (Zhang et al., 2005). In this study, OPR2 was only responsive to pathogen attack at the later time point when disease was already established (data not shown). Collectively, the described expression of PR genes as well as OPR2 suggests that SA, a major signal in SAR responses triggered by pathogen infection, is most likely not a major player in T. virens-mediated systemic resistance in maize.
We further tested the expression of the JA/ET-responsive and -biosynthetic genes in T. virens-mediated resistance responses. Expression of PAL has been reported to be activated by the JA/ET signaling pathway (Diallinas and Kanellis, 1994; Kato et al., 2000; Shoresh et al., 2005). PAL is the first enzyme in the phenylpropanoid biosynthetic pathway, which provides precursors for the formation of an array of antimicrobial compounds (Dixon et al., 2002). We have previously shown that treatment of cotton cotyledons with Sm1 elicitor resulted in increased levels of autofluorescence (Djonovic et al., 2006a), which is usually associated with accumulation and oxidation of phenolic compounds such as phytoalexins and lignin (Nicholson and Hammerschmidt, 1992; Heath, 2000). Howell et al. (2000) found that cotton radicles treated with T. virens or T. virens protein fractions exhibited elevated levels of the biosynthesis of sesquiterpenoid phytoalexins. The consistent pattern of up-regulated PAL expression in T. virens wild type- and overexpression strain-inoculated maize (both locally and systemically) and the similar basal expression in nontreated and deletion strain-treated plants illustrate the importance of this gene and the corresponding pathway for T. virens-Sm1-mediated defense responses in maize.
Existing evidence has implicated 13-LOX-derived oxylipins, including jasmonates and GLVs, in plant resistance mechanisms against diverse pathogens and pests. Both jasmonates produced by the AOS branch of the LOX pathway, and GLVs, the derivatives of the HPL branch, have potent signaling activities that regulate expression of numerous defense-related and developmental genes (Bate and Rothstein, 1998; Birkett et al., 2000; Turner et al., 2002). Our study shows that AOS may be of relevance to T. virens-Sm1-mediated defense responses in maize because AOS transcripts were up-regulated by wild-type and overexpression strains of T. virens but remained low in nontreated and deletion strain-treated plants. Recently, it was shown that overexpression of the AOS gene in rice increased endogenous JA levels, followed by enhanced resistance to fungal infection (Mei et al., 2006). Another gene from the JA biosynthetic pathway, OPR7, which was previously shown to be highly induced in maize by JA/ET treatments but not by SA (Zhang et al., 2005), was also induced by T. virens wild-type and overexpression strains. Interestingly, a gene encoding the putative GLV-producing enzyme HPL exhibited the highest level of expression in plants induced by a SM1-overexpression strain. Similar to this finding, the T. asperellum-mediated systemic disease protection of cucumber plants from a bacterial leaf pathogen was also associated with increased levels of HPL transcripts (Yedidia et al., 2003). Priming of bean plants with nonpathogenic Pseudomonas putida reduced the symptoms caused by the pathogen Botrytis cinerea and was accompanied by stimulation of the LOX pathway that may have resulted in higher levels of endogenous GLVs (Ongena et al., 2004). Indeed, overexpression of the HPL gene in Arabidopsis resulted in enhanced resistance against B. cinerea (Shiojiri et al., 2006). Additionally, very recently, GLVs were shown to play an important “within-plant” role in defense signaling (Heil and Silva Bueno, 2007).
Taken together, the gene expression data suggest that T. virens induces systemic resistance in maize by an ISR-like mechanism involving JA/ET/GLV-mediated pathways rather than a SAR-like mechanism that requires SA.
Sm1 Is Expressed in Maize Roots during Interaction with T. virens But Does Not Affect Root Colonization
The role of hydrophobins in several mutualistic symbioses has been reported previously (Honegger, 1991; Tagu et al., 1996; Scherrer et al., 2000). Recently, a hydrophobin gene, TasHyd1, was detected during cucumber root colonization by T. asperellum. Transformants disrupted in this gene were severely impaired in root attachment and colonization (Viterbo and Chet, 2006). Thus, we examined strains disrupted in SM1 to test their ability to colonize maize roots. We did not detect any difference among wild-type and transformant strains as all colonized maize roots at very high efficiency. For example, in the hydroponic system, after 2 d of growth, more than 94% root fragments were colonized by all strains of T. virens (Table I). Additionally, GFP-transformed wild type and deletion transformants showed a similar pattern of colonization on maize roots (Fig. 7).
While the colonization of roots by T. virens does not yield the elaborate hyphal structures seen with ectomycorrhizal fungi or the level of penetration as with arbuscular mycorrhizal fungi, an intimate association is formed that involves a signaling process (Yedidia et al., 2003). Here, we have shown that several maize defense genes were affected by the presence of Sm1. Maize seedlings grown with transformants that fail to produce Sm1 express these same defense genes at levels similar to the non-Trichoderma-treated plants. The involvement of Sm1 in the induction of defense responses was further supported by demonstrating the expression of SM1 in roots colonized by Trichoderma before and after pathogen challenge. This finding indicated again that besides being an elicitor, this protein may have some further signaling role in plant-fungal interaction as proposed earlier (Djonovic et al., 2006a). For example, the hydrophobins HydPt and HydPt-1, produced during the early stages of Pisolithus-Eucalyptus ectomycorrhizae formation, have been proposed to be involved in the adhesion of the mycelia to the root surface (Tagu et al., 1996). In T. asperellum, hydrophobin TasHyd1 was proposed to provide a protection to the hyphae from plant defense compounds during the first steps of colonization of the intercellular spaces of the plant roots (Viterbo and Chet, 2006). We are currently examining other potential roles of Sm1 by identifying plant proteins that interact with Sm1.
Possible Role of Sm1 in T. virens Physiology
Sm1 is a member of a new cerato-platanin (Pazzagli et al., 1999) family of hydrophobin-like, small (approximately 150 amino acids) secreted proteins, mainly associated with toxicity and infection processes, and produced by plant and human fungal pathogens. In contrast to these reported functions of cerato-platanin members, we have demonstrated that Sm1, produced by a beneficial microorganism T. virens, does not have toxic activity and instead is an effective elicitor of systemic resistance (Djonovic et al., 2006a). Very little is known about the role of these proteins in fungal development and physiology (Boddi et al., 2004). When a SM1 homolog from Leptosphaeria maculans, SP1, was disrupted, it was shown not to be crucial for pathogenicity; however, the effect of the SP1 deletion for fungal development has not been reported (Wilson et al., 2002). Here, we show that neither deletion nor overexpression of SM1 altered the fungal phenotypic traits (i.e. colony appearance and pigmentation during sporulation, germination, growth, hydrophobicity of mycelia or conidia, and mycoparasitic characteristics such as coiling and production of gliotoxin) we examined. Other roles in the development of T. virens may yet be realized for Sm1 as cerato-platanin, a homolog of Sm1, has been shown to be located in the cell walls of ascospores, hyphae, and conidia, and the authors have proposed that this protein may have some new structural functions for Ceratocystis fimbriata (Boddi et al., 2004). Further studies are in progress to address the role of Sm1 in fungal development using SM1∷GFP fusion strains (C.M. Kenerley, unpublished data).
In summary, this study provides evidence that, similar to dicotyledonous plants, colonization of maize roots by T. virens induces ISR-like systemic protection associated with notable induction of JA- and GLV-biosynthetic genes. We also demonstrated that the activity of a functional Sm1 elicitor was required for T. virens-mediated ISR. The fact that overexpression of this gene led to enhanced levels of systemic protection suggests that genetic manipulation of fungal biocontrol agents or plants to express this elicitor may lead to improved crop resistance against pests.
MATERIALS AND METHODS
Fungal and Plant Materials
Two strains of Trichoderma virens were used in this study, a wild-type strain, Gv29-8, and an Arg auxotrophic strain, Tv10.4, as the recipient for fungal transformation (Baek and Kenerley, 1998). An isolate of the foliar pathogen of maize (Zea mays), Colletotrichum graminicola, was kindly provided by Dr. S. Sukno (Texas A&M University). The isolate of Rhizopus oryzae were kindly provided by Dr. C. Howell (Southern Plains Agricultural Research Center, U.S. Department of Agriculture Agricultural Research Service, College Station, TX). The strains were routinely maintained on PDA (Difco) with C. graminicola grown under constant light. For screening of transformants, VMS and PDA were used.
Maize (inbred line B73) seedlings used in this study were grown in a hydroponic system (Djonovic et al., 2006a) or in the soil-less growth medium Metro-Mix 366 (Scotts). The disease resistance bioassay was performed with maize plants grown in a growth chamber at 25°C and with a 14-h photoperiod and 60% humidity.
Construction of SM1-Disruption and -Overexpression Vectors
To obtain the flanking regions of the SM1 ORF for construction of deletion strains, a clone with a high-molecular-mass insert was isolated from T. virens BAC library as described previously (Djonovic et al., 2006a). This BAC clone was further digested with several restriction enzymes and the fragments subcloned into pBluescript II SK (±) to obtain the following vectors: pSZD14 (a 5.6-kb SacI subclone), pSZD15 (a 1-kb HindIII subclone), and pSZD21 (a 7.5-kb ClaI subclone) that were used for sequencing and further analysis. Nucleotide sequencing was performed by a primer-walking strategy. All sequencing reactions were performed at the Gene Technologies Laboratory (Texas A&M University). DNA sequences were analyzed by DNA Strider 1.2 and Sequencher 4.1 (Gene Code Corporation). The QIAprep spin miniprep kit (Qiagen) was used for plasmid DNA purification.
The SM1-disruption vector (pSZD25) was constructed by replacing a 483-bp ORF with a 3.0-kb SmaI/EcoRV fragment of the T. virens ARG2 gene (Baek and Kenerley, 1998), which served as a selectable marker for transformation. The resulting vector contained 2.1-kb and 1.7-kb noncoding upstream and downstream SM1 sequences from pSZD21 flanking the selectable ARG2 gene. The 5.6-kb PstI/XbaI deletion cassette was isolated from pSZD25 and used for fungal transformation. An overexpression vector (pSZD26) was constructed for constitutive overproduction of the SM1 gene by placing a 1-kb HindIII insert from pSDZ15 subclone containing the SM1 ORF between the promoter and the terminator regions of the T. virens GPD (glyceraldehyde-3-P dehydrogenase) gene (Xu et al., 1996). To obtain overexpression strains, Tv10.4 was cotransformed with pSZD26 and pJMB4, which contain ARG2 gene as a selectable marker.
Transformation and Screening of Transformants
Stable prototrophic transformants were selected by consecutive transfer of single colonies to VMS, PDA, and VMS (Baek and Kenerley, 1998). Screening of potential deletion and overexpression transformants was first performed by PCR. For deletion transformants, SM1-specific primers SmF and SmR (Djonovic et al., 2006a) were used to amplify the 264-bp PCR product from the wild-type genomic DNA. PCR amplification of the SM1 fragment comprised 28 cycles (each cycle: 30 s at 94°C, 20 s at 55°C, and 20 s at 72°C). Strains that yielded no PCR product were further screened by Southern analysis. For overexpression strains, forward (SmF) and reverse (R50) 5′-TACAGACAATGATTCATG-3′ primers were designed to amplify a 1-kb fragment from the SM1 ORF and a GPD terminator region from the overexpression cassette. Strains that yielded the 1-kb PCR product were selected for further analysis by Southern blotting. PCR amplification comprised 28 cycles (each cycle: 30 s at 94°C, 20 s at 40°C, and 30 s at 72°C). PCR was performed according to manufacturer's instructions using the Invitrogen Taq DNA polymerase kit.
Northern Analysis of Transformants
Conidia of 7-d-old T. virens wild type or transformants cultured on PDA were used to inoculate VMS (for deletion strains) or GYEC (15 g of Glc, 3 g of yeast extract, 5 g of casein hydrolysate per liter; for overexpression strains) liquid medium (Thomas and Kenerley, 1989) to a final concentration of 106 spores/mL. All cultures were incubated on an orbital shaker (130 rpm) at room temperature. After 5 and 3 d of growth for deletion and overexpression strains, respectively, the mycelia were harvested and rinsed thoroughly with sterile water. Total RNA was extracted from the harvested mycelia as described previously (Djonovic et al., 2006a). Fifteen micrograms for deletion stains and 10 μg for overexpression strains of total RNA per sample was denaturated, resolved in 1.5% agarose formaldehyde gel, transferred to a Hydrobond-N+ nylon membrane (Amersham Biosciences), and hybridized overnight at 42°C using Ultrahyb (Ambion). The 264-bp (SmF-SmR) PCR product was used as SM1 probe. The blots were reprobed with a 550-bp PCR product from the actin (ACT) gene (Djonovic et al., 2006b) as positive control for even loading.
SDS-PAGE and Western Analysis of Transformants
Fungal CFs were obtained by inoculating 200 mL of VMS with a conidial suspension of the appropriate fungal strain to a final concentration of 106/mL conidia. Following incubation on a rotary shaker at 130 rpm for 5 d at 23°C, CFs were successively filtered through a 10-μm NITEX nylon cloth (TETKO) and a 0.45-μm filter (Fisher Scientific). Proteins in the CFs were precipitated by 80% ammonium sulfate (Fisher Scientific). Pellets were resuspended in small amounts of 10 mm Tris, pH 7.8, and dialyzed against the same buffer (10-kD mwco; Pierce). Protein concentrations were determined by Bio-Rad Bradford microassay using bovine serum albumin as a standard. Protein extracts were subjected to SDS-PAGE following silver or Coomassie Brilliant Blue R-250 staining for protein visualization. Prestained SDS-PAGE broad-range molecular mass standards (Bio-Rad) or Kaleidoscope polypeptide molecular mass standards (Bio-Rad) were used for molecular mass determination.
Protein extracts (obtained as described above) were electrophoresed on SDS-PAGE gels and electroblotted to a nitrocellulose membrane (Osmonics). Sm1 protein was detected using Sm1 polyclonal antibodies (dilution 1:1,000) in a standard western-blot procedure (Sambrook et al., 1989). SDS-PAGE and western analysis for comparison of Sm1 expression in wild-type, overexpression, and deletion strains hydroponically grown with maize seedlings was performed as we have reported previously (Djonovic et al., 2006a).
Phenotypic Analysis of Transformants
Cultures of selected transformants were compared with the wild-type strain for colony morphology and radial growth. Agar plugs from actively growing colonies were inoculated in the center of VMS, PDA, or WA plates. Plates were visually inspected for production of aerial hyphae, and color and morphology of the colony. Hyphal extension was recorded at 24 and 48 h of growth at 27°C. The border of the hyphal extension was marked each day, and each plate was photographed and the surface area of growth for each day determined using ImageJ software (http://rsb.info.nih.gov/ij/). Each treatment contained four repetitions and each experiment was repeated at least twice. Data were analyzed by ANOVA and Fisher's PLSD test (P < 0.05; Statview Version 5.0.1; SAS Institute).
For germination study, conidial suspensions (1 × 106 /mL) of each strain were plated onto PDA and incubated for 10 d at 27°C. Sterile glass slides were coated with approximately 1 mL of PDA. Conidia were gently removed from the plates and spread onto the coated slides at a concentration of 103 per slide. The slides were incubated for 12 h at 27°C in the dark in moist chambers. The number of germinated conidia was determined by observing 100 conidia along random transects on the slides. Each strain was replicated three times and the experiment repeated at least twice.
Two methods were used to assay hydrophobicity of the mutants and the wild type. In the first assay, each strain was assayed on PDA solid medium at 2 d prior to sporulation. Water drops (20 μL) were placed on the surface and digitally photographed immediately. The shape of the drop and the angle of contact of the drop with the colony provide an indication of the hydrophobicity of the surface (van der Mei et al., 1991). The second method involves separation of conidia into aqueous and organic phases to assess hydrophobicity (Whiteford and Spanu, 2001). Briefly, the conidia of each strain are diluted in phosphate buffered saline to obtain an optical density (OD) of about 0.7 at 420 nm. A 750-μL aliquot of this dilution is added to 250 μL of hexadecane. This solution is vortexed for 1 min, allowed to settle for 5 min, 200 μL is added to a 96-well plate, and the optical density recorded with a microwell plate reader. Increasing amounts of triton were added to the conidia prior to the addition of the hexadecane to lower the affinity of the conidia for the aqueous-organic interphase.
For experiment to detect the coiling phase of mycoparasitism, glass slides were coated with VMS by dipping the slides into the molten agar. An agar plug (3 mm in diameter) of the selected strain of T. virens was added to one side of the coated slide and incubated at 27°C for 36 h. The isolate of R. oryzae was then placed (agar plugs) opposite the hyphae of T. virens (1–1.5 cm) and the slides further incubated at 27°C. After 24 to 48 h, the slides were microscopically examined (see section “Microscopy and Imaging”) for evidence of mycoparasitism (coiling) at the zones of interaction.
Infection of Trichoderma-Treated Maize Plants with C. graminicola
Cultures of C. graminicola for inoculation were grown for 14 d on PDA at room temperature under constant light. Conidia were scraped from plates, filtered through Miracloth (Calbiochem), and washed three times in distilled water, followed by centrifugation for 1 min at 10,000 rpm. Conidia were counted by using a hemacytometer, spore suspension was adjusted to 6.5 × 104 conidia/mL, and Tween 20 was added to a final concentration of 0.005%.
Inoculation of maize seeds with the wild type, KO26, KO46, OE38, or OE39 was performed by first coating the seeds with a latex sticker (Phoplex AC-33; Rohm and Haas), followed by the addition of fine powder of chlamydospore preparations (Weaver and Kenerley, 2005). Control (nontreated seeds) and fungus-coated seeds were planted in containers (3.81 × 20.9 cm; Stuewe & Sons) containing a soil-less mix (Metro-Mix 366) and incubated in a growth chamber (EGC) at 25°C, with a 14-h photoperiod and 60% humidity, for 2 weeks.
Fourteen-day-old plants (at the V4 developmental stage) were inoculated with C. graminicola by placing the plants in trays (78.7 × 63.5 × 6.9 cm; Molded Fiber Glass Tray Company) and taping the leaves onto moist paper towels in the bottom of the trays. The third leaf from all plants was inoculated with six droplets (10 μL), each containing 650 conidia, placed on the adaxial side, away from the midvein of the leaf. The trays were sealed with plastic wrap to maintain the moisture and incubated for 24 h. After incubation, the plants were left for 3 to 4 h for droplets to dry, and the location of each droplet was marked to allow for later identification. Then, the plants were gently untaped, returned to controlled growth conditions, and monitored for the appearance of symptoms. Following a 4-d incubation period, inoculated leaves were scanned and percentage of the leaf area with symptomatic lesions was determined using ImageJ software. Each treatment, consisting of six spots per leaf per one plant, was replicated four times, and the experiment was repeated three times. Data were analyzed by ANOVA and Fisher's PLSD test (P < 0.05; Statview Version 5.0.1; SAS Institute).
T. virens-Root Colonization Studies
Plants were removed from the hydroponic system and the roots were excised from the plant. The entire root system was placed in 100 mL of 1% sodium hexametaphosphate and 0.1% Tergitol P10 (Sigma Aldrich) and shaken for 45 min at 100 rpm on an orbital shaker. The roots were then removed, rinsed twice in sterile water, and then shaken for 45 min in 100 mL of sterile water. Root systems were then spread apart in a petri dish (150 mm diameter) containing sterile water and harvested (excised with a scalpel) for plating on GVSM (Park et al., 1992). Excised root fragments were blotted dry on sterile filter paper before placement in GVSM. After 48 h of incubation at 27°C, the total length and length colonized by T. virens were measured. As these roots were extensively colonized, the length of the root from which numerous coalescing colonies emerged was used as a measure of colonization.
After 14 d of growth in soil, plants were removed from containers, shoots excised from the root systems, root systems extensively washed under tap water and collected on a sieve. The same washing protocol was followed as cited above. Roots were excised and plated on GVSM. Over a 48 h period of incubation, colonies of T. virens growing from the root fragments onto GVSM were assessed. Root fragments were measured and the data are expressed as colonies/cm of root length.
GFP-Tagged T. virens Strain Construction
T. virens wild-type and KO25 strains expressing GFP were constructed by cotransformation (Djonovic et al., 2007) using the plasmids pTEFEGFP and pCSN44. pTEFEGFP contains the GFP coding sequence under the action of the Aureobasidium pullulans translation elongation factor promoter and the Aspergillus awamori glucoamylase terminator (Vanden Wymelenberg et al., 1997). Plasmid pCSN44 containing HPH (Staben et al., 1989) was used as a selectable marker for cotransformation procedure.
Stable cotransformants were selected on PDA plates supplemented with hygromycin (Dave et al., 1994). Resistant colonies were screened for the presence of green fluorescence and the insertion of pTEFEGFP was further analyzed by Southern-blot analysis. Genomic DNA was prepared and digested using BamHI or HindIII restriction endonucleases. DNA digests were electrophoresed in 1% agarose gels, blotted onto nylon membranes, and probed with GFP or HPH gene-specific probes. The growth rate and colony morphology of selected cotransformants were similar to the original strains as judged by growth comparison on PDA, VMS, and WA plates.
Expression Analysis of Maize Defense-Related Genes
Expression of defense-related genes was analyzed in maize seedlings (roots and leaves) grown hydroponically without or with wild-type, SM1-deletion, and SM1-overexpression strains (as described above). In the experiment with maize seeds coated with wild-type strain, two deletion strains, and two overexpression strains (described in section above) and challenged with C. graminicola, the seedlings were grown in soil. Plants were harvested after 14 d of growth with Trichoderma, 1 d after incubation with the pathogen, and 4 d after inoculation when the symptoms were evaluated. All harvested samples were immediately frozen in liquid nitrogen. Total RNA from maize was extracted using TRI reagent (Molecular Research Center) according to the manufacturer's protocols.
Expression of plant defense-related genes in this study was assessed by sqRT-PCR. Sequences for gene-specific primer pairs were obtained from published studies (ZmOPR2, ZmOPR7, and ZmOPR8 [Zhang et al., 2005]; ZmPAL and ZmHPL [Farag et al., 2005]; AOS [He et al., 2005]; and ZmLOX10 [Nemchenko et al., 2006]), whereas primer pairs for PR genes were designed based on sequences available in the GenBank database, namely, ZmPR1 (U82200) and ZmPR5 (U82201; Morris et al., 1998; Gao et al., 2007). The forward and reverse primers were: 5′-AAC AAT GGC ACC GAG GCT AGC GT-3′and 5′-GTA TGC ATG ACA GTC TAG TAG GG-3′, respectively, for ZmPR1, and 5′-TAG CTC TAT AGC TCG AGT ATT GCT-3′ and 5′-TCA CTA GCC CAT GCA TGC AGA GC-3′ for ZmPR5. Maize GAPc (Farag et al., 2005) was used as an internal control. Extracted RNA was DNase treated and cleaned using a DNA-free kit (Ambion). Total RNA (2.5 μg) was reverse transcribed with a first-strand cDNA synthesis kit (G.E. Healthcare) using the random hexamer pd(N)6 as a primer. To define the optimal number of PCR cycles for linear amplification of each gene, a range of PCR amplifications was performed. Subsequently, PCR products were electrophoresed, stained with ethidium bromide, and band signals quantified by Scion image software (http://www.scioncorp.com/). PCR amplification of PAL and GAPc gene fragments in both hydroponic and soil-grown maize seedlings experiments comprised 25 cycles (each cycle: 30 s at 94°C, 30 s at 58°C, and 30 s at 72°C) for leaf RNA and 23 cycles for PAL root RNA. To detect the expression of genes that were expressed at very low levels (i.e. PR1, PR5, OPR2, and LOX10), the number of PCR cycles for those genes was increased by an additional one to three cycles. PCR amplification for PR1, PR5, OPR7, OPR8, and HPL gene fragments comprised 28 cycles (each cycle: 30 s at 94°C, 30 s at 58°C, and 30 s at 72°C); OPR2: 28 cycles (each cycle: 30 s at 94°C, 30 s at 53°C, and 1.3 min at 72°C); AOS: 28 cycles (each cycle: 30 s at 94°C, 30 s at 56°C, and 28 s at 72°C); and LOX10: 29 cycles (each cycle: 30 s at 94°C, 30 s at 50°C, and 15 s at 72°C). The negative control for RT-PCR was DNase-treated and cleaned RNA reactions without reverse transcription. PCR was performed according to the manufacturer's instructions using the Invitrogen Taq DNA polymerase kit. PCR products were electrophoresed on 2% agarose gels, and band intensities compared within each experiment after ethidium bromide staining and intensities of bands were quantified using Scion Image software.
In Planta SM1 mRNA Detection
To detect SM1 mRNA in roots of maize plants inoculated with T. virens wild-type or mutant strains, total RNA from roots was extracted and used for RT-PCR experiments. After harvesting, the roots were washed with tergitol-metaphosphate solution as described above and total RNA was extracted. Reverse transcription reactions were performed using 2.5 μg of total RNA. The reactions for SM1 and ACT genes from T. virens were carried out for 40 cycles of 94°C (30 s), 58°C (30 s), and 72°C (30 s), plus a single final step at 72°C for 5 min, and using the primers 5′GACACTGGTGAGACAAGCAC3′ (forward) and 5′TTAGAGACCGCAGTTCTTAACAG3′ (reverse) for the SM1 gene or 5′GTATCATGATCGGTATGGGTCAGAA3′ (forward) and 5′TAGAAGGTGTGGTGCCAGATCTT3′ (reverse) for the ACT gene. PCR reactions for GAPc were conducted on the conditions mentioned above. PCR products were separated on 2% agarose gels, ethidium bromide stained, and visualized under UV light. Maize defense gene expression was profiled from two independent experiments. As the results from the two experiments were similar, the data from one trial are presented here.
Microscopy and Imaging
Microscopy of coiling experiments was carried out with an Olympus BX60 microscope and 100× magnification. The images were captured using the Q-Free Olympus camera and processed using Adobe Photoshop imaging software. Fluorescence microscopy was performed with an Olympus BX51 fluorescence microscope. Excitation and emission wavelengths of 470 to 490 and 510 to 550 nm, respectively, and 200× magnification were used. Images were recorded using an Olympus DP70 camera and processed with DPController 1.1.165 software.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Coiling capability of wild-type, SM1-deletion, and SM1-overexpression strains.
Supplemental Table S1. Growth analysis of agar plates inoculated with wild-type, SM1-deletion, or SM1-overexpression strains.
Supplementary Material
Acknowledgments
We thank Dr. Serenella Sukno for sharing the protocol for Colletotrichum inoculation of maize; Dr. Brain Shaw for expert assistance in fluorescence microscopy; Dr. Daniel Cullen for providing the GFP vector and Dr. Deborah Bell-Pedersen for providing the hygromycin vector; Dr. Xiquan Gao and Dr. Andriy Nemchenko for providing the primers for maize gene expression; and Gloria Vittone for assistance with plant bioassays and hydrophobicity assay.
This work was supported by grants from the U.S. Department of Agriculture National Research Initiative (2003–35316–13861) and the National Science Foundation (IOB0445650) to C.M.K., and a fellowship to S.D. from the Storkan-Hanes-McCaslin Foundation.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Charles M. Kenerley (c-kenerley@tamu.edu).
The online version of this article contains Web-only data.
References
- Agrawal GK, Jwa NS, Rakwal R (2000) A novel rice (Oryza sativa L.) acidic PR1 gene highly responsive to cut, phytohormones, and protein phosphatase inhibitors. Biochem Biophys Res Commun 274 157–165 [DOI] [PubMed] [Google Scholar]
- Baek JM, Kenerley CM (1998) The arg2 gene of Trichoderma virens: cloning and development of a homologous transformation system. Fungal Genet Biol 23 34–44 [DOI] [PubMed] [Google Scholar]
- Bailey BA, Korcak RF, Anderson JD (1992) Alterations in Nicotiana tabacum L. cv xanthi cell membrane function following treatment with an ethylene biosynthesis-inducing endoxylanase. Plant Physiol 100 749–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bais HP, Park SW, Weir TL, Callaway RM, Vivanco JM (2004) How plants communicate using the underground information superhighway. Trends Plant Sci 9 26–32 [DOI] [PubMed] [Google Scholar]
- Bate NJ, Rothstein SJ (1998) C6-volatiles derived from the lipoxygenase pathway induce a subset of defense-related genes. Plant J 16 561–569 [DOI] [PubMed] [Google Scholar]
- Benitez T, Rincon AM, Limon MC, Codon AC (2004) Biocontrol mechanisms of Trichoderma strains. Int Microbiol 7 249–260 [PubMed] [Google Scholar]
- Birkett MA, Campbell CA, Chamberlain K, Guerrieri E, Hick AJ, Martin JL, Matthes M, Napier JA, Pettersson J, Pickett JA, et al (2000) New roles for cis-jasmone as an insect semiochemical and in plant defense. Proc Natl Acad Sci USA 97 9329–9334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boddi S, Comparini C, Calamassi R, Pazzagli L, Cappugi G, Scala A (2004) Cerato-platanin protein is located in the cell walls of ascospores, conidia and hyphae of Ceratocystis fimbriata f. sp. platani. FEMS Microbiol Lett 233 341–346 [DOI] [PubMed] [Google Scholar]
- Calderon AA, Zapata JM, Munoz R, Pedreno MA, Barcelo AR (1993) Resveratrol production as a part of the hypersensitive-like response of grapevine cells to an elicitor from Trichoderma viride. New Phytol 124 455–463 [Google Scholar]
- Chern M, Fitzgerald HA, Canlas PE, Navarre DA, Ronald PC (2005) Overexpression of a rice NPR1 homolog leads to constitutive activation of defense response and hypersensitivity to light. Mol Plant Microbe Interact 18 511–520 [DOI] [PubMed] [Google Scholar]
- Conrath U, Beckers GJ, Flors V, Garcia-Agustin P, Jakab G, Mauch F, Newman MA, Pieterse CM, Poinssot B, Pozo MJ, et al (2006) Priming: getting ready for battle. Mol Plant Microbe Interact 19 1062–1071 [DOI] [PubMed] [Google Scholar]
- Dave KI, Lauriano C, Xu B, Wild JR, Kenerley CM (1994) Expression of organophosphate hydrolase in the filamentous fungus Gliocladium virens. Appl Microbiol Biotechnol 41 352–358 [DOI] [PubMed] [Google Scholar]
- Diallinas G, Kanellis AK (1994) A phenylalanine ammonia-lyase gene from melon fruit: cDNA cloning, sequence and expression in response to development and wounding. Plant Mol Biol 26 473–479 [DOI] [PubMed] [Google Scholar]
- Dixon RA, Achnine L, Kota P, Liu CJ, Reddy MSS, Wang LJ (2002) The phenylpropanoid pathway and plant defence—a genomics perspective. Mol Plant Pathol 3 371–390 [DOI] [PubMed] [Google Scholar]
- Djonovic S, Pozo MJ, Dangott LJ, Howell CR, Kenerley CM (2006. a) Sm1, a proteinaceous elicitor secreted by the biocontrol fungus Trichoderma virens induces plant defense responses and systemic resistance. Mol Plant Microbe Interact 19 838–853 [DOI] [PubMed] [Google Scholar]
- Djonovic S, Pozo MJ, Kenerley CM (2006. b) Tv-bgn3, a beta-1,6-glucanase from the biocontrol fungus Trichoderma virens is involved in mycoparasitism and control of Pythium ultimum. Appl Environ Microbiol 72 7661–7670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djonovic S, Vittone G, Mendoza-Herrera A, Kenerley C (2007) Enhanced biocontrol activity of Trichoderma virens transformants constitutively co-expressing β-1,3- and β-1,6-glucanase genes. Mol Plant Pathol 8 469–480 [DOI] [PubMed] [Google Scholar]
- Dong X (2001) Genetic dissection of systemic acquired resistance. Curr Opin Plant Biol 4 309–314 [DOI] [PubMed] [Google Scholar]
- Dong X (2004) NPR1, all things considered. Curr Opin Plant Biol 7 547–552 [DOI] [PubMed] [Google Scholar]
- Duplessis S, Courty PE, Tagu D, Martin F (2005) Transcript patterns associated with ectomycorrhiza development in Eucalyptus globulus and Pisolithus microcarpus. New Phytol 165 599–611 [DOI] [PubMed] [Google Scholar]
- Durrant WE, Dong X (2004) Systemic acquired resistance. Annu Rev Phytopathol 42 185–209 [DOI] [PubMed] [Google Scholar]
- Farag MA, Fokar M, Zhang HA, Allen RD, Pare PW (2005) (Z)-3-Hexenol induces defense genes and downstream metabolites in maize. Planta 220 900–909 [DOI] [PubMed] [Google Scholar]
- Feussner I, Wasternack C (2002) The lipoxygenase pathway. Annu Rev Plant Biol 53 275–297 [DOI] [PubMed] [Google Scholar]
- Furman-Matarasso N, Cohen E, Du Q, Chejanovsky N, Hanania U, Avni A (1999) A point mutation in the ethylene-inducing xylanase elicitor inhibits the β-1-4-endoxylanase activity but not the elicitation activity. Plant Physiol 121 345–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Starr J, Göbel C, Engelberth J, Feussner I, Tumlinson J, Kolomiets M (2007) Maize 9-lipoxygenase ZmLOX3 controls development, root-specific expression of defense genes and resistance to root-knot nematodes. Mol Plant Microbe Interact (in press) [DOI] [PubMed]
- Glazebrook J (2005) Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu Rev Phytopathol 43 205–227 [DOI] [PubMed] [Google Scholar]
- Glazebrook J, Chen WJ, Estes B, Chang HS, Nawrath C, Metraux JP, Zhu T, Katagiri F (2003) Topology of the network integrating salicylate and jasmonate signal transduction derived from global expression phenotyping. Plant J 34 217–228 [DOI] [PubMed] [Google Scholar]
- Gozzo F (2003) Systemic acquired resistance in crop protection: from nature to a chemical approach. J Agric Food Chem 51 4487–4503 [DOI] [PubMed] [Google Scholar]
- Hanson LE, Howell CR (2004) Elicitors of plant defense responses from biocontrol strains of Trichoderma virens. Phytopathology 94 171–176 [DOI] [PubMed] [Google Scholar]
- Harman GE, Howell CR, Viterbo A, Chet I, Lorito M (2004. a) Trichoderma species—opportunistic, avirulent plant symbionts. Nat Rev Microbiol 2 43–56 [DOI] [PubMed] [Google Scholar]
- Harman GE, Petzoldt R, Comis A, Chen J (2004. b) Interactions between Trichoderma harzianum strain T22 and maize inbred line Mo17 and effects of these interactions on diseases caused by Pythium ultimum and Colletotrichum graminicola. Phytopathology 94 147–153 [DOI] [PubMed] [Google Scholar]
- He GZ, Tarui Y, Iino M (2005) A novel receptor kinase involved in jasmonate-mediated wound and phytochrome signaling in maize coleoptiles. Plant Cell Physiol 46 870–883 [DOI] [PubMed] [Google Scholar]
- Heath MC (2000) Hypersensitive response-related death. Plant Mol Biol 44 321–334 [DOI] [PubMed] [Google Scholar]
- Heil M, Bostock RM (2002) Induced systemic resistance (ISR) against pathogens in the context of induced plant defences. Ann Bot (Lond) 89 503–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heil M, Silva Bueno JC (2007) Within-plant signaling by volatiles leads to induction and priming of an indirect plant defense in nature. Proc Natl Acad Sci USA 104 5467–5472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honegger R (1991) Functional aspects of the lichen symbiosis. Annu Rev Plant Physiol Plant Mol Biol 42 553–578 [Google Scholar]
- Howell CR, Hanson LE, Stipanovic RD, Puckhaber LS (2000) Induction of terpenoid synthesis in cotton roots and control of Rhizoctonia solani by seed treatment with Trichoderma virens. Phytopathology 90 248–252 [DOI] [PubMed] [Google Scholar]
- Hwang J, Benson DM (2003) Expression of induced systemic resistance in poinsettia cuttings against Rhizoctonia stem rot by treatment of stock plants with binucleate Rhizoctonia. Biol Control 27 73–80 [Google Scholar]
- Kato M, Hayakawa Y, Hyodo H, Ikoma Y, Yano M (2000) Wound-induced ethylene synthesis and expression and formation of 1-aminocyclopropane-1-carboxylate (ACC) synthase, ACC oxidase, phenylalanine ammonia-lyase, and peroxidase in wounded mesocarp tissue of Cucurbita maxima. Plant Cell Physiol 41 440–447 [DOI] [PubMed] [Google Scholar]
- Kogel KH, Beckhove U, Dreschers J, Munch S, Romme Y (1994) Acquired resistance in barley (the resistance mechanism induced by 2,6-dichloroisonicotinic acid is a phenocopy of a genetically based mechanism governing race-specific powdery mildew resistance). Plant Physiol 106 1269–1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Floch G, Benhamou N, Mamaca E, Salerno MI, Tirilly Y, Rey P (2005) Characterisation of the early events in atypical tomato root colonisation by a biocontrol agent, Pythium oligandrum. Plant Physiol Biochem 43 1–11 [DOI] [PubMed] [Google Scholar]
- Matsui K (2006) Green leaf volatiles: hydroperoxide lyase pathway of oxylipin metabolism. Curr Opin Plant Biol 9 274–280 [DOI] [PubMed] [Google Scholar]
- Mei C, Qi M, Sheng G, Yang Y (2006) Inducible overexpression of a rice allene oxide synthase gene increases the endogenous jasmonic acid level, PR gene expression, and host resistance to fungal infection. Mol Plant Microbe Interact 19 1127–1137 [DOI] [PubMed] [Google Scholar]
- Mittler R, Vanderauwera S, Gollery M, Van Breusegem F (2004) Reactive oxygen gene network of plants. Trends Plant Sci 9 490–498 [DOI] [PubMed] [Google Scholar]
- Morris SW, Vernooij B, Titatarn S, Starrett M, Thomas S, Wiltse CC, Frederiksen RA, Bhandhufalck A, Hulbert S, Uknes S (1998) Induced resistance responses in maize. Mol Plant Microbe Interact 11 643–658 [DOI] [PubMed] [Google Scholar]
- Nemchenko A, Kunze S, Feussner I, Kolomiets M (2006) Duplicate maize 13-lipoxygenase genes are differentially regulated by circadian rhythm, cold stress, wounding, pathogen infection, and hormonal treatments. J Exp Bot 57 3767–3779 [DOI] [PubMed] [Google Scholar]
- Nicholson RL, Hammerschmidt R (1992) Phenolic compounds and their role in disease resistance. Annu Rev Phytopathol 30 369–389 [Google Scholar]
- Nimchuk Z, Eulgem T, Holt BF III, Dangl JL (2003) Recognition and response in the plant immune system. Annu Rev Genet 37 579–609 [DOI] [PubMed] [Google Scholar]
- Ongena M, Duby F, Rossignol F, Fauconnier ML, Dommes J, Thonart P (2004) Stimulation of the lipoxygenase pathway is associated with systemic resistance induced in bean by a nonpathogenic Pseudomonas strain. Mol Plant Microbe Interact 17 1009–1018 [DOI] [PubMed] [Google Scholar]
- Park YH, Kenerley CM, Stack JP (1992) Inoculum dynamics of Gliocladium virens associated with roots of cotton seedlings. Microb Ecol 23 169–179 [DOI] [PubMed] [Google Scholar]
- Paul ND, Hatcher PE, Taylor JE (2000) Coping with multiple enemies: an integration of molecular and ecological perspectives. Trends Plant Sci 5 220–225 [DOI] [PubMed] [Google Scholar]
- Pazzagli L, Cappugi G, Manao G, Camici G, Santini A, Scala A (1999) Purification, characterization, and amino acid sequence of cerato-platanin, a new phytotoxic protein from Ceratocystis fimbriata f. sp. platani. J Biol Chem 274 24959–24964 [DOI] [PubMed] [Google Scholar]
- Pieterse CM, van Loon LC (1999) Salicylic acid-independent plant defence pathways. Trends Plant Sci 4 52–58 [DOI] [PubMed] [Google Scholar]
- Pieterse CMJ, Van Pelt JA, Verhagen BWM, Ton J, Van Wees SCM, Leon-Kloosterziel KM, Van Loon LC (2003) Induced systemic resistance by plant growth-promoting rhizobacteria. Symbiosis 35 39–54 [Google Scholar]
- Pozo MJ, Van Loon LC, Pieterse CMJ (2005) Jasmonates—signals in plant-microbe interactions. J Plant Growth Regul 23 211–222 [Google Scholar]
- Rakwal R, Komatsu S (2000) Role of jasmonate in the rice (Oryza sativa L.) self-defense mechanism using proteome analysis. Electrophoresis 21 2492–2500 [DOI] [PubMed] [Google Scholar]
- Rotblat B, Enshell-Seijffers D, Gershoni JM, Schuster S, Avni A (2002) Identification of an essential component of the elicitation active site of the EIX protein elicitor. Plant J 32 1049–1055 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Schaffrath U, Freydl E, Dudler R (1997) Evidence for different signaling pathways activated by inducers of acquired resistance in wheat. Mol Plant Microbe Interact 10 779–783 [Google Scholar]
- Schenk PM, Kazan K, Wilson I, Anderson JP, Richmond T, Somerville SC, Manners JM (2000) Coordinated plant defense responses in Arabidopsis revealed by microarray analysis. Proc Natl Acad Sci USA 97 11655–11660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherrer S, De Vries OMH, Dudler R, Wessels JGH, Honegger R (2000) Interfacial self-assembly of fungal hydrophobins of the lichen-forming ascomycetes Xanthoria parietina and X. ectaneoides. Fungal Genet Biol 30 81–93 [DOI] [PubMed] [Google Scholar]
- Schweizer P, Schlagenhauf E, Schaffrath U, Dudler R (1999) Different patterns of host genes are induced in rice by Pseudomonas syringae, a biological inducer of resistance, and the chemical inducer benzothiadiazole (BTH). Eur J Plant Pathol 105 659–665 [Google Scholar]
- Shiojiri K, Kishimoto K, Ozawa R, Kugimiya S, Urashimo S, Arimura G, Horiuchi J, Nishioka T, Matsui K, Takabayashi J (2006) Changing green leaf volatile biosynthesis in plants: an approach for improving plant resistance against both herbivores and pathogens. Proc Natl Acad Sci USA 103 16672–16676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoresh M, Gal-On A, Leibman D, Chet I (2006) Characterization of a mitogen-activated protein kinase gene from cucumber required for Trichoderma-conferred plant resistance. Plant Physiol 142 1169–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoresh M, Yedidia I, Chet I (2005) Involvement of jasmonic acid/ethylene signaling pathway in the systemic resistance induced in cucumber by Trichoderma asperellum T203. Phytopathology 95 76–84 [DOI] [PubMed] [Google Scholar]
- Staben C, Jensen B, Singer M, Pollock J, Schechtman M, Kinsey J, Selker E (1989) Use of a bacterial hygromycin B resistance gene as a dominant selectable marker in Neurospora crassa transformation. Fungal Genet Newsl 36 79–81 [Google Scholar]
- Tagu D, Lapeyrie F, Martin F (2002) The ectomycorrhizal symbiosis: genetics and development. Plant Soil 244 97–105 [Google Scholar]
- Tagu D, Nasse B, Martin F (1996) Cloning and characterization of hydrophobins-encoding cDNAs from the ectomycorrhizal Basidiomycete Pisolithus tinctorius. Gene 168 93–97 [DOI] [PubMed] [Google Scholar]
- Thomas MD, Kenerley CM (1989) Transformation of the mycoparasite Gliocladium. Curr Genet 15 415–420 [Google Scholar]
- Ton J, Van Pelt JA, Van Loon LC, Pieterse CMJ (2002) Differential effectiveness of salicylate-dependent and jasmonate/ethylene-dependent induced resistance in Arabidopsis. Mol Plant Microbe Interact 15 27–34 [DOI] [PubMed] [Google Scholar]
- Turner JG, Ellis C, Devoto A (2002) The jasmonate signal pathway. Plant Cell (Suppl) 14 S153–S164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Mei HC, Rosenberg M, Busscher HJ (1991) Assessment of microbial cell surface hydrophobicity. In N Mozes, PS Handley, HJ Busscher, PG Rouxhet, eds, Microbial Cell Surface Analysis. VCH, New York, pp 265–287
- van Loon LC, Bakker PA, Pieterse CM (1998) Systemic resistance induced by rhizosphere bacteria. Annu Rev Phytopathol 36 453–483 [DOI] [PubMed] [Google Scholar]
- Van Loon LC, Van Strien EA (1999) The families of pathogenesis-related proteins, their activities, and comparative analysis of PR-1 type proteins. Physiol Mol Plant Pathol 55 85–97 [Google Scholar]
- Vanden Wymelenberg AJ, Cullen D, Spear RN, Schoenike B, Andrews JH (1997) Expression of green fluorescent protein in Aureobasidium pullulans and quantification of the fungus on leaf surfaces. Biotechniques 23 686–690 [DOI] [PubMed] [Google Scholar]
- Veronese P, Ruiz MT, Coca MA, Hernandez-Lopez A, Lee H, Ibeas JI, Damsz B, Pardo JM, Hasegawa PM, Bressan RA, et al (2003) In defense against pathogens. Both plant sentinels and foot soldiers need to know the enemy. Plant Physiol 131 1580–1590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viterbo A, Chet I (2006) TasHyd1, a new hydrophobin gene from the biocontrol agent Trichoderma asperellum, is involved in plant root colonization. Mol Plant Pathol 7 249–258 [DOI] [PubMed] [Google Scholar]
- Viterbo A, Harel M, Chet I (2004) Isolation of two aspartyl proteases from Trichoderma asperellum expressed during colonization of cucumber roots. FEMS Microbiol Lett 238 151–158 [DOI] [PubMed] [Google Scholar]
- Viterbo A, Harel M, Horwitz BA, Chet I, Mukherjee PK (2005) Trichoderma mitogen-activated protein kinase signaling is involved in induction of plant systemic resistance. Appl Environ Microbiol 71 6241–6246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel HJ (1956) A convenient growth medium for Neurospora (Medium N). Microbiol Genet Bull 13 42–43 [Google Scholar]
- Waller F, Achatz B, Baltruschat H, Fodor J, Becker K, Fischer M, Heier T, Huckelhoven R, Neumann C, von Wettstein D, et al (2005) The endophytic fungus Piriformospora indica reprograms barley to salt-stress tolerance, disease resistance, and higher yield. Proc Natl Acad Sci USA 102 13386–13391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver M, Kenerley C (2005) Density independent population dynamics by Trichoderma virens in soil and defined substrates. Biocontrol Sci Technol 15 847–857 [Google Scholar]
- Whiteford JR, Spanu PD (2001) The hydrophobin HCf-1 of Cladosporium fulvum is required for efficient water-mediated dispersal of conidia. Fungal Genet Biol 32 159–168 [DOI] [PubMed] [Google Scholar]
- Wilson LM, Idnurm A, Howlett BJ (2002) Characterization of a gene (sp1) encoding a secreted protein from Leptosphaeria maculans, the blackleg pathogen of Brassica napus. Mol Plant Pathol 3 487–493 [DOI] [PubMed] [Google Scholar]
- Xu BW, Wild JR, Kenerley CM (1996) Enhanced expression of a bacterial gene for pesticide degradation in a common soil fungus. J Ferment Bioeng 81 473–481 [Google Scholar]
- Yedidia I, Benhamou N, Chet I (1999) Induction of defense responses in cucumber plants (Cucumis sativus L.) by the biocontrol agent Trichoderma harzianum. Appl Environ Microbiol 65 1061–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yedidia I, Benhamou N, Kapulnik Y, Chet I (2000) Induction and accumulation of PR proteins activity during early stages of root colonization by the mycoparasite Trichoderma harzianum strain T-203. Plant Physiol Biochem 38 863–873 [Google Scholar]
- Yedidia I, Shoresh M, Kerem Z, Benhamou N, Kapulnik Y, Chet I (2003) Concomitant induction of systemic resistance to Pseudomonas spingae pv. lachrymans in cucumber by Trichoderma asperellum (T-203) and accumulation of phytoalexins. Appl Environ Microbiol 69 7343–7353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Simmons C, Yalpani N, Crane V, Wilkinson H, Kolomiets M (2005) Genomic analysis of the 12-oxo-phytodienoic acid reductase gene family of Zea mays. Plant Mol Biol 59 323–343 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.