Abstract
Plant thioredoxins (TRXs) are involved in redox regulation of a wide variety processes and usually exhibit organ specificity. We report strong evidence that chloroplastic TRXs are localized in heterotrophic tissues and suggest some ways in which they might participate in several metabolic and developmental processes. The promoter regions of the chloroplastic f and m1 TRX genes were isolated from a pea (Pisum sativum) plant genomic bank. Histochemical staining for β-glucuronidase (GUS) in transgenic homozygous Arabidopsis (Arabidopsis thaliana) plants showed preferential expression of the 444-bp PsTRXf1 promoter in early seedlings, stems, leaves, and roots, as well as in flowers, stigma, pollen grains, and filaments. GUS activity under the control of the 1,874-bp PsTRXm1 promoter was restricted to the leaves, roots, seeds, and flowers. To gain insight into the translational regulation of these genes, a series of deletions of 5′ elements in both TRX promoters were analyzed. The results revealed that a 126-bp construct of the PsTRXf2 promoter was unable to reproduce the expression pattern observed with the full promoter. The differences in expression and tissue specificity between PsTRXm1 and the deleted promoters PsTRXm2 and PsTRXm3 suggest the existence of upstream positive or negative regulatory regions that affect tissue specificity, sucrose metabolism, and light regulation. PsTRXm1 expression is finely regulated by light and possibly by other metabolic factors. In situ hybridization experiments confirmed new localizations of these chloroplastic TRX transcripts in vascular tissues and flowers, and therefore suggest possible new functions in heterotrophic tissues related to cell division, germination, and plant reproduction.
Much progress has been made in recent years in discovering specific functions and protein targets of plant thioredoxins (TRXs). However, different TRXs can be active within the same organelle and may have redundant functions or act on different protein targets. This situation makes it difficult to determine their specificity and raises questions concerning their individuality (Balmer et al., 2004). The discovery of at least 20 different TRXs during sequencing of the Arabidopsis (Arabidopsis thaliana) genome has shown both the organization of this multigene family and the evolutionary pathways of the genes involved to be complex (Meyer et al., 2005).
TRXs are small (12–14 kD) proteins with a conserved redox-active site (WCXPC) involved in oxidoreductase activity with thiol-disulfide interchange reactions in all organisms (Holmgren, 1985). In its reduced form, TRX can function as a hydrogen donor or as a regulatory factor for various target proteins, such as metabolic enzymes, redox proteins, transcription factors, or mitogen-activated protein kinases (for review, see Arner and Holmgren, 2000; Schürmann and Jacquot, 2000). The bridge formed between two Cys of the TRX protein is reduced by ferredoxin via ferredoxin-TRX reductase (FTR) in the chloroplast, whereas cytosolic TRX is reduced by NADPH via NADPH-TRX reductase in other organelles and different parts of the plant.
Plant TRXs are organized in different groups depending on their localization. The fully sequenced Arabidopsis genome disclosed nine TRXs named h-type (heterotrophic) proteins localized in the cytosol, two mitochondrial TRX o proteins, and several chloroplastic TRXs encoded by four TRX m genes, two TRX f genes, one TRX x gene, and two TRX y genes. The specificity of TRX h is currently under review. However, several studies based on yeast complementation, proteomics, and protein target identification methods have indicated important new functions for these cytosolic TRXs and suggested a role for these proteins in different stress situations (Mouaheb et al., 1998; Wong et al., 2004; Traverso et al., 2007). Several potential targets for TRX o proteins have emerged recently, but no specific function has yet been discovered for them (Balmer et al., 2004). Among the chloroplastic TRX proteins, the f and m isoforms are the best described, and their function in carbon metabolism of the chloroplast has been well defined (Crawford et al., 1979; Mestres-Ortega and Meyer, 1999; Meyer et al., 2002; Collin et al., 2003, 2004). The chloroplastic TRX m is of prokaryote origin, whereas the chloroplastic TRX f and cytosolic TRX h share an intron position and seem to have a common eukaryote ancestor (Sahrawy et al., 1996).
Research in pea (Pisum sativum) plants on the roles of TRXs has given rise to some confusion. To date, one TRX f and two TRX m (m1 and m2) have been found in the chloroplast (López-Jaramillo et al., 1997; Pagano et al., 2000), and four TRX h are localized in the cytosol (Montrichard et al., 2003; Traverso et al., 2007). Both TRX f and TRX m regulate key enzymes that play an essential role in carbon fixation and sugar biosynthesis during photosynthesis. TRX f specifically activates photosynthetic Fru-1,6-bisphosphatase (FBPase), whereas malate dehydrogenase (NADP-MDH) is the target of TRX m (Bassham and Krause, 1969; López-Jaramillo et al., 1997). However, the distinction between the reactivity of the two chloroplastic isoforms with their target enzymes has not always been clear. Under certain assay conditions, TRX f can also activate NADP-MDH (Hodges et al., 1994; Geck et al., 1996), whereas FBPase can also be the target of TRX m (López-Jaramillo et al., 1997), but the efficiency of activation of the two targets differs. In consonance with their functions and localization, the biosynthesis of chloroplastic TRXs is induced by light (Carrasco et al., 1992).
A few years ago, it was reported that TRX f accumulates in roots as both a protein and a transcript (Pagano et al., 2000). This was the first finding of chloroplastic TRXs in a nonphotosynthetic organ. Recently, a TRX m isoform and the complete ferredoxin-TRX system were identified in the amyloplast of wheat (Triticum aestivum) endosperm (Balmer et al., 2006a). Several potential protein targets for chloroplastic TRX m have been identified with proteomics-based methods, but no new functions have been determined. Some studies have raised the possibility that chloroplastic TRXs participate in metabolic processes other than those related to photosynthesis in almost all stages of plant development. Despite extensive studies of the structure, function, and regulation of plant TRXs, however, little is known about the roles of plant TRXs. Few studies have focused on the search for new localization sites of chloroplastic TRXs or on the discovery of new roles in heterotrophic tissues.
Because the information provided by promoter regions could yield evidence of the sites of action and roles of different TRXs, we isolated the pea chloroplastic TRX f and TRX m promoters and analyzed their pattern of expression using the GUS reporter gene in Arabidopsis plants, which confirmed the presence of these TRXs in nonphotosynthetic organs. Database analysis of the chloroplastic TRX promoter regions identified several important cis-regulatory sequences related to tissue localization, light, and metabolite regulation, and serial deletions of these regulatory regions revealed that chloroplastic TRX m was finely regulated by the surrounding tissue and by exposure to light. We also developed an in situ hybridization method to detect chloroplastic TRX mRNA expression in any plant organ and structure.
We discuss how these results shed light on new sites of chloroplastic TRX action and analyze the regulation of these proteins in light of the cis-acting elements found in the promoter sequences. These considerations allow us to suggest possible new functions for chloroplastic TRXs in addition to their already well-known role in regulating carbon metabolism enzymes in the chloroplasts of green tissues.
RESULTS
Cloning and Structural Analysis of Pea TRX f and TRX m Promoters Revealed cis-Elements Related to Localization and Metabolic Regulation
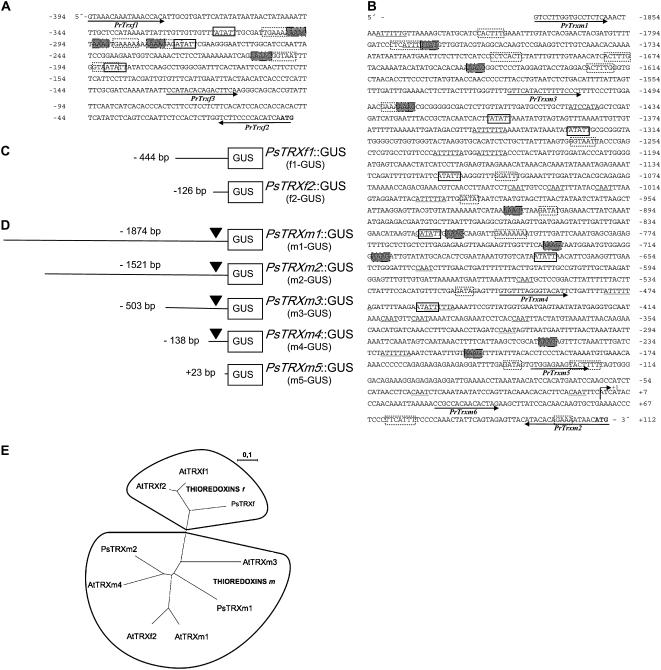
We used the PCR-walking technique to isolate from several pea genomic DNA libraries two promoters corresponding to TRX f and TRX m1 genes. To avoid confusion, TRX m1 is designated TRX m throughout the text. The promoter of the TRX f gene is 444 bp upstream from the translation start site, whereas the promoter of the TRX m gene is 1,973 bp upstream from the ATG. The nucleotide sequences of the promoter fragment from PsTRX f and the 5′ flanking sequence from PsTRX m are presented in Figure 1, A and B. Although the TRX f promoter is shorter than expected, it contains the principal cis-acting elements that allow sufficient expression. RACE assays mapped the transcription start site to an A nucleotide 112 nucleotides upstream from the translation initiation codon of the TRX m gene (indicated as +1 in Fig. 1B). Despite several attempts, we were not able to determine the transcription start position of the TRX f gene. Bioinformatics analysis was undertaken to identify conserved motifs found in other eukaryotic promoters and to find putative cis-elements that might be essential in the regulation of TRX f and TRX m gene expression (Fig. 1). No typical TATA box was identified; however, for both TRXs, a putative TATA box was located at −31 bp from the transcription start site for the TRX m gene and −128 bp from the ATG for TRX f.
Figure 1.
Sequence of the 5′ upstream region of the pea TRX f (A) and TRX m (B) genes, deleted promoter constructions of TRX f (C) and TRX m (D), and phylogenetic analysis (E). Nucleotides are numbered relative to the transcription start site indicated as +1 for the TRX m promoter and the ATG for TRX f. Putative cis-elements found in the database are indicated in boxes. Root-specific tissues are in white boxes, carbon metabolism-related elements are in shaded boxes, light-responsive elements are in dashed boxes, and elements related to seed development are underlined. C, Schema of the serial deletions of the promoter of the TRX f gene fused to the GUS reporter gene. D, Schema of the serial deletions of the promoter of the TRX m gene fused to the GUS reporter gene. E, ClustalX phylogenetic tree for chloroplastic TRX f and TRX m from pea (Ps) and Arabidopsis (At). Accession numbers are (in parentheses): PsTRXf (CAA45098), PsTRXm1 (CAA53900), PsTRXm2 (CAC69854), AtTRXf1 (NP_186922), AtTRXf2 (NP_197144), AtTRXm1 (NP_849585), AtTRXm2 (NP_192261), AtTRXm3 (NP_179159), and AtTRXm4 (NP_188155).
We classified the potential regulatory elements in the pea TRX f promoter into three categories: light-, tissue-, and carbon metabolism-dependent elements (Fig. 1A). Among the light-dependent elements were several GATA motif sequences and Ibox, as well as a GAAAAA motif at position −315, a GTAATA element at position −274, and a GTAATT sequence at position −190 (Lam and Chua, 1989; Gilmartin et al., 1990). Root tissue-specific elements (ATATT) were also found (Elmayan and Tepfer, 1995). The promoter sequence also contained four Dof-like elements (AAAG) related to the promoter of genes that encode enzymes involved in carbon metabolism (Yanagisawa, 2000).
Among the putative regulatory elements, the TRX m promoter comprised at least 16 different light-dependent elements, such as GATA, GT1, and Ibox (Fig. 1B). Several root tissue-specific cis-acting sequences (ATATT) were localized, as well as three CAAT box-like sequences at positions −7, −43, and −323 that have been found in promoters of genes encoding seed storage protein (Shirsat et al., 1989). The ATTTTTA sequence is related to the regulation of plant development and seed storage proteins. Finally, a number of sequences that bind Dof elements (AAAG) conserved in promoters of genes involved in sugar biosynthesis were identified in the TRX m promoter.
Our comparison of the two TRX promoters (Fig. 1) showed that, despite their different sizes, both contained similar cis-acting elements that determine their specific localization in the plant, their regulation by light, and their function during carbon metabolism, aspects that we considered important for the study of chloroplastic TRXs' localization and regulation. The bioinformatics analysis revealed, in addition, elements involved in processes of embryo and seed formation, protein storage, circadian clock, pollen localization, and hormonal regulation.
Because it was impossible to transform pea plants, we obtained Arabidopsis transgenic plants that carried the pea TRX f and TRX m promoters. Although this heterologous system is widely used in plant molecular biology, we decided to compare chloroplastic TRXs in both plant species at the protein and promoter sequence level to validate our results. A comparative phylogeny analysis of chloroplastic TRXs f and m from pea and Arabidopsis showed that PsTRX f was grouped with AtTRX f1 and AtTRX f2, whereas the four Arabidopsis TRXs m were positioned together with PsTRX m1 and PsTRX m2 (Fig. 1E). These data confirmed the high similarity between both f-type and m-type TRX species: PsTRX f had 86% identity with AtTRX f1 and 90% identity with AtTRX f2, whereas PsTRX m1 shared 82%, 82%, 71%, and 82% identity with AtTRX m1, AtTRX m2, AtTRX m3, and AtTRX m4, respectively.
In The Arabidopsis Information Resource (TAIR) database, we examined the promoter regions of the two TRXs f and four TRXs m of Arabidopsis: AtTRX m1 (AT1G03680), AtTRX m2 (AT4G03520), AtTRX m3 (AT2G15570), AtTRX m4 (AT3G15360), AtTRX f1 (AT3G02730), and AtTRX f2 (AT5G16400). Bioinformatics analysis detected similar regulatory boxes in pea TRX f and TRX m promoters that were related to light, tissue, and carbon metabolism.
Promoter∷GUS Fusion in Transgenic Arabidopsis Plants Revealed Expression in Green and Nonphotosynthetic Tissues
To examine the contribution of the 5′ region of chloroplastic TRX genes to the regulation of expression, different extensions of 5′ flanking sequences of TRX f and TRX m were transcriptionally fused to the uidA (GUS) reporter gene via the pBI101 vector, and designated PsTRXf1∷GUS (f1-GUS) or PsTRXf2∷GUS (f2-GUS) with the TRX f promoter, and PsTRXm1∷GUS (m1-GUS), PsTRXm2∷GUS (m2-GUS), PsTRXm3∷GUS (m3-GUS), PsTRXm4∷GUS (m4-GUS), or PsTRXm5∷GUS (m5-GUS) with the TRX m promoter (Fig. 1, C and D). After transformation of Arabidopsis plantlets and generation of homozygous lines that expressed the promoter TRX∷GUS fusion genes, we examined plantlets for tissue-specific GUS activity. Enzyme activity was not detected in control plants transformed with an empty pBI101 vector.
PsTRX f∷GUS Fusion
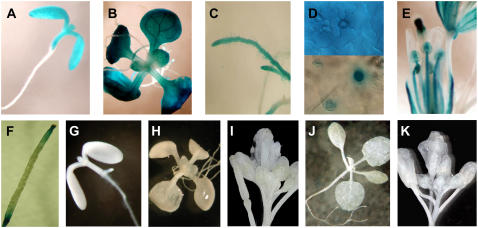
At early stages of germination in plants transformed with PsTRXf1, GUS was strongly expressed mainly in the cotyledons, hypocotyl, and meristematic ring (Fig. 2A). After 7 to 10 d of growth, GUS activity appeared in leaves, leaf primordia (Fig. 2B), and root apical meristems (Fig. 2C). Staining was also observed in the stomata and at the base of the trichome (Fig. 2D). In fully expanded plantlets (20 to 30 d old), flowers of transgenic plants that expressed the TRX f promoter∷GUS fusion showed histochemical staining exclusively in anthers, pollen grains, filaments, and stigmas (Fig. 2E), and at the top and base of the siliques (Fig. 2F). Vascular tissues showed a weak, but clear, signal in leaves and sepals. The deletion of 267 bp of the promoter induced the complete loss of GUS expression in cotyledons (Fig. 2G) and fully developed plants (Fig. 2, H and I). Transgenic plants containing the empty control vector pBI101 showed no staining (Fig. 2, J and K).
Figure 2.
Histochemical localization of GUS expression under the control of the PsTRXf1∷GUS promoter (A–F) and PsTRXf2∷GUS deleted promoter (G–I) in transgenic Arabidopsis plants. A, Seven-day-old seedling. B, Fifteen-day-old plant grown in tissue culture, showing strong GUS expression in the cotyledons, hypocotyls, and leaves. C, GUS expression in the root apical meristem of a 15-d-old plant. D, Stomata showing expression in the guard cells. E, Flowers of 21- to 30-d-old plant showing stained pollen grains, filaments, and stigmas. F, Siliques showing GUS activity at the top and bottom of developing siliques. G (seedling), H (15-d-old plant), and I (flower), Deleted PsTRX f2∷GUS promoter (f2-GUS) showing complete loss of expression. J and K, Vector pBI101 in control transgenic plants. J, Fifteen-day-old plant. K, Twenty-one-day-old plant.
PsTRX m∷GUS Fusion
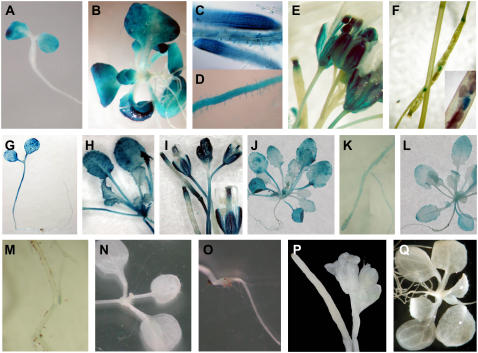
In the early stages of germination of plants transformed with the TRX m promoter∷GUS fusion, staining was restricted to cotyledons (Fig. 3A). A few days later, transgenic plants displayed prominent GUS activity in leaves (Fig. 3B), as well as in regions of intense cell division, such as the apical meristem and procambium region, and in the root apical meristem and epidermis, including root hairs (Fig. 3, C and D). In flowers, we observed a specific signal in pollen grains and filaments as well as in stigmas (Fig. 3E). After 3 weeks of development, GUS staining appeared in the intersection of the secondary and primary stems and at the end of the siliques. Seeds inside siliques showed a weak, but localized, signal (Fig. 3F). Deletion of 1,877 bp of the TRX m promoter resulted in complete loss of GUS accumulation in all organs of PsTRXm5∷GUS transgenic Arabidopsis plantlets (Fig. 3, N–P). The sequence comprising the region from position −138 to +23 bp of the 259-bp fragment (PsTRXm4∷GUS) was sufficient to reproduce most features of the expression pattern seen with the complete promoter sequence and to maintain basal expression in cotyledons, hypocotyls, roots, and trichomes of seedlings. As plants matured, however, GUS expression became weaker in leaves (Fig. 3L) and almost disappeared in roots (Fig. 3M). In contrast to the results with the PsTRXm1∷GUS fusion, plants transformed with the PsTRXm2∷GUS and PsTRXm3∷GUS fusions showed additional staining in the hypocotyl (Fig. 3G). Leaves of the plantlets accumulated high levels of GUS (Fig. 3, H and J), whereas expression in the roots decreased significantly (Fig. 3K). In flowers of PsTRXm2∷GUS, PsTRXm3∷GUS, and PsTRXm4∷GUS plants, weak staining was detected in anther filaments, but staining was intense in stigmas (Fig. 3I). These results were confirmed in two independent reporter lines. The intensity of GUS activity in the leaves of transgenic plants containing the TRX f or TRX m promoter decreased as plant senescence progressed. Transgenic plants containing the empty control vector pBI101 showed no staining (Fig. 3Q).
Figure 3.
Histochemical localization of GUS activity under the control of the TRX m promoter. A to F, GUS expression in transgenic Arabidopsis plants carrying the PsTRXm1∷GUS fusion construct. A, GUS staining localized in the cotyledons of 7-d-old seedlings. B and C, Leaves and roots from a 15-d-old plantlet. C and D, GUS activity localized in root apical meristem and root hairs of 15-d-old plantlet. E, GUS staining localized in flowers of a 21-d-old plant. F, GUS expression in seeds in the silique. G to I, GUS expression under the deleted promoter PsTRXm2∷GUS. G, Additional staining in the hypocotyl of 7-d-old seedlings. H, GUS activity throughout the aerial part of a 15-d-old plant. I, Weaker GUS signals are visible in flowers, with the exception of strong staining in the stigma of a 21- to 30-d-old plant. J and K, Aerial part and root of a 15-d-old plant showing GUS expression under the deleted promoter PsTRXm3∷GUS. The pattern of expression is similar to that detected in PsTRXm2∷GUS transgenic plants. L and M, Aerial part and root of a 15-d-old plant, showing GUS activity under the deleted promoter PsTRXm4∷GUS. The signal in this transgenic line is similar to that found with the previous deleted construct. Assays with the PsTRXm5∷GUS fusion yielded no expression, as shown in N (10-d-old seedlings), O (root of a 15-d-old plant), and P (flowers of a 21- to 30-d-old plant). Q, Vector pBI101 in control transgenic plants.
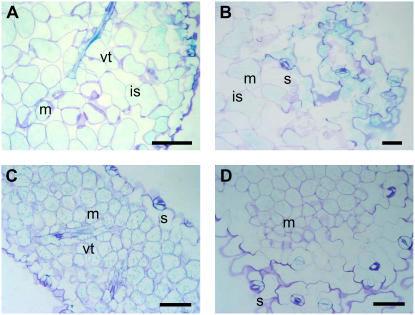
Figure 4 shows a cross section of a GUS-stained Arabidopsis leaf expressing the PsTRXm1∷GUS and PsTRXf1∷GUS promoters. TRX f (Fig. 4, A and B) and TRX m (Fig. 4, C and D) were located in the chloroplast of mesophyll cells and in stomata. Surprisingly, a strong signal was also seen in the vascular tissue of the leaf and in the tracheids, as observed in transgenic plants.
Figure 4.
Semithin cross sections of leaves from GUS-stained plants transformed with f1-GUS (A and B) and m1-GUS (C and D). Bars = 50 μm. GUS staining is observed in mesophyll cells, vascular tissue, and stomata. m, Mesophyll; s, stomata; vt, vascular tissue; is, intercellular spaces.
Semiquantitative PCR Expression of GUS Revealed Light-Dependent Regulation
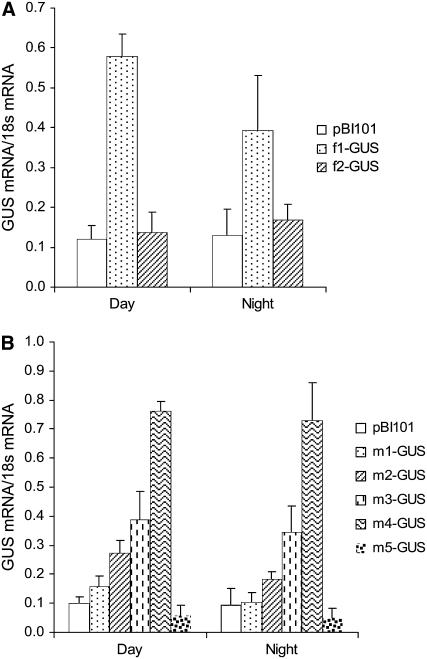
In Arabidopsis plants carrying the PsTRXf1∷GUS construct and subjected to a 12-h-light/12-h-dark photoperiod, GUS RNA expression displayed a peak at the middle of the day that decayed by 33% during the dark period in comparison to the light period (Fig. 5A). This light-dependent expression disappeared in plants carrying the shorter fragment of the TRX f promoter (PsTRXf2∷GUS), and the level of messenger was similar during the day and night phases (Fig. 5A). We also found high GUS expression in transgenic plants under the control of the PsTRXm1∷GUS promoter; expression was most intense after 8 h of light and decreased during the dark period by 31% (Fig. 5B). The pattern of expression was similar in plants bearing the PsTRXm2∷GUS: high expression during the light period, which decreased by 33% during the night. Surprisingly, the PsTRXm3∷GUS and PsTRXm4∷GUS promoters yielded similar levels of expression during the day and the night, with decreases during the dark period of only 10% and 4%, respectively, in comparison to the light period. Light-induced expression disappeared completely in plants with the PsTRXm5∷GUS promoter. Unexpectedly, with the exception of PsTRXm5∷GUS, serial deletions of the TRX m promoter induced a progressive increase in GUS mRNA expression in response to light. In plants with the PsTRXm4∷GUS promoter, mRNA expression was up to 5-fold as high as the expression of PsTRXm1∷GUS mRNA.
Figure 5.
Light/dark regulation of GUS mRNA expression under TRX f and TRX m promoters. Transgenic plants carrying complete promoter constructs (f1-GUS or m1-GUS) or a truncated promoter (f2-GUS, m2-GUS, m3-GUS, m4-GUS, or m5-GUS) were subjected to a 12-h-day/12-h-night photoperiod, and mRNA levels of the GUS-encoding gene were determined in leaves by semiquantitative RT-PCR as described in “Materials and Methods.” A, GUS expression under the TRX f promoter. B, GUS expression under the TRX m promoter. Samples were harvested at midday and midnight from 20-d-old plants. Values are the average of three determinations of two cDNA preparations.
In Situ Hybridization Revealed New Sites of Action in Reproductive Organs and Vascular Tissues
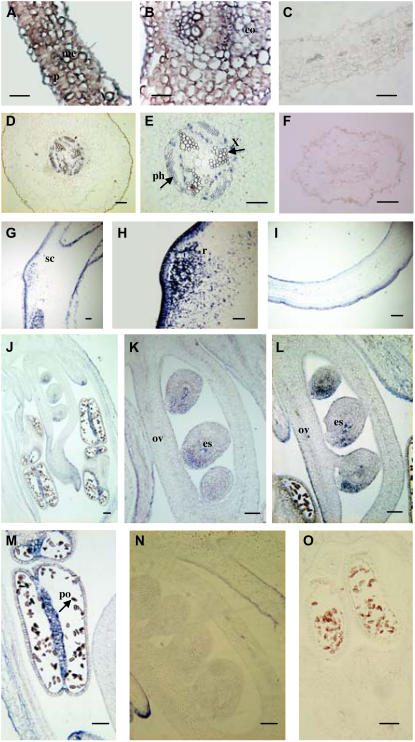
To determine the patterns of expression of the TRX f and TRX m pea chloroplastic genes, we conducted in situ hybridization with a digoxigenin-labeled antisense RNA probe specific to each of the chloroplastic TRX cDNA sequences in different pea tissues. Leaf cross sections revealed TRX f and TRX m mRNA throughout the parenchyma and mesophyll layers (Figs. 6A and 7A). However, TRX f expression was more intense in collenchyma cells and regions adjacent to the leaf midvein and around the xylem vessel and phloem sieve tube, as observed in cross sections of stained leaves (Fig. 6, A and B). In contrast, intense TRX m signals were detected in the mesophyll cells of the leaf (Fig. 7B) and low expression was detected in the parenchyma (Fig. 7A). Again, we were able to detect a strong signal around the vascular tissues, phloem, and xylem, and neither of the TRXs seemed to be expressed in the epidermis. In root cross sections, we found localized expression of TRX f in the vascular tissue (Fig. 6D) and vascular tissue triarchs (Fig. 6E). When the same tissues were assayed with sense probes, no specific reaction was detected (Fig. 6, C and F). Pea seeds also accumulated TRX f messenger in the outer membrane of the coat seed and in a region of active cell division that likely corresponded to radicle protrusion (Fig. 6, G and H). A strong signal was detected in ovules, pollen grains, and connective tissue when flower longitudinal sections were hybridized with a TRX f antisense probe (Fig. 6J). However, no clearly defined structure could be identified inside the pollen (Fig. 6M). Two intense signals appeared inside the ovule. One area of expression was localized in the middle of the embryonic sac, an area corresponding to the endosperm tissue which, soon after fertilization, starts to undergo cell division at a high rate to provide nutrients. The second area was localized in the external part of the ovule and might be related to the developing integument, which grows to cover the nucella (Fig. 6, K and L). Sense experiments confirmed that the probes used were specific for their hybridization (Fig. 6, I, N, and O).
Figure 6.
Localization of TRX f mRNA in pea plants by in situ hybridization with the TRX f antisense probe. Bars = 100 μm. A and B, Cross and longitudinal sections of a leaf. C, Leaf cross section hybridized with sense probe. D, Root cross section. E, Root cross section at higher magnification showing positive reaction around the vascular area. F, Root cross section hybridized with the sense probe. G and H, Longitudinal section of pollen grains. I, Control sample of pollen grains hybridized with sense probe. J to M, Longitudinal section of flowers at different stages. J, Developing flower. K and L, Details of developing embryos. M, Pollen grains in the anthers. N and O, Control samples of flower sections hybridized with the sense probe. me, Mesophyll; p, parenchyma; co, collenchyma; x, xylem; ph, phloem; sc, seed coat; r, radicle; ov, ovary; es, embryonic sac; po, pollen.
Figure 7.
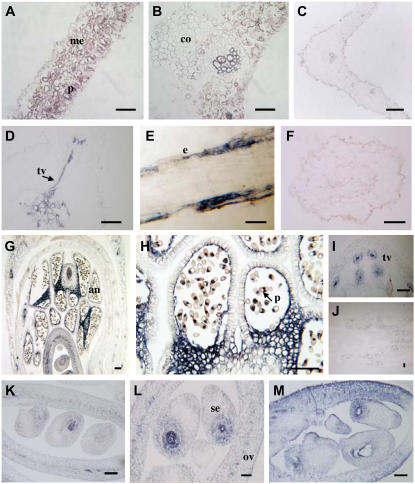
Localization of TRX m in pea plants by in situ hybridization with the TRX m antisense probe. Bars = 100 μm. A and B, Cross and longitudinal sections of a leaf. C, Leaf section hybridized with the sense probe. D, Cross section of a root. E, Longitudinal section of a root. F, Sense control of TRX m mRNA in roots. G, Longitudinal section of a flower. H, Pollen grain. I, Vascular tissue of the pedicel. J, Longitudinal section of a flower showing the sense control. K to M, Higher magnifications of longitudinal sections of ovaries showing details of developing embryos. me, Mesophyll; p, parenchyma; co, collenchyma; vt, vascular tissue; e, epidermis; al, anther locus; po, pollen; ov, ovary; es, embryonic sac.
In root cross sections, a strong signal was evident in vascular tissues (Fig. 7D). By comparing the results with those in sense probe-treated tissue (Fig. 7F), we detected TRX m mainly in the xylem, with only weak reactivity in the phloem. This pattern of expression was also seen in the secondary vascular tissue toward lateral roots. Observations in longitudinal sections of roots confirmed that TRX m was expressed in the epidermis of the differentiation zone (Fig. 7E). We also observed that TRX m was widely expressed in reproductive organs (Fig. 7G) and was prominent in pollen grains (Fig. 7H), tapetal cells of the anthers, ovules, and connective tissue. The two signals in pollen grains appeared to be localized in the generative cell and tube cell, the latter of which is involved in the process of forming the pollen tube for pollination.
In later stages of flowering, the signal in pollen grains disappeared and the embryo displayed a strong reaction localized in the middle of the embryonic sac, a site of intense cell division during endosperm synthesis (Fig. 7, H, K, and L). The vascular tissue of reproductive organs also expressed TRX m, as seen in the pedicel (Fig. 7I). No signal was observed upon hybridization with the TRX m sense probe in leaves (Fig. 7C), roots (Fig. 7F), or flowers (Fig. 7J).
Pea Tissue Expression Detected FTR and TRX f and TRX m in Different Tissues
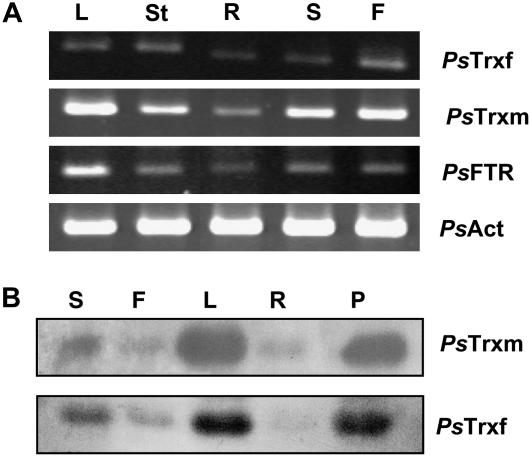
The cDNA obtained by reverse transcription (RT) of RNA extracted from pea leaves, stems, roots, seeds, and flowers was subjected to PCR with specific oligonucleotides corresponding to the TRX f, TRX m, and FTR genes. As expected, all three genes were expressed in leaves and stems (Fig. 8A; Table I); additionally, a significant level of mRNA expression was also detected in seeds as well as in nonphotosynthetic tissues, such as roots, and in flowers. The TRX m gene was highly expressed in leaves, seeds, and flowers. Expression of TRX f was clearly positive, but comparatively weaker, and appeared mainly in stems, leaves, and flowers. The level of FTR mRNA was higher in leaves than in other tissues. In all cases, we detected the presence of the three genes in roots and flowers, indicating a possible role for chloroplastic TRXs in heterotrophic tissues.
Figure 8.
A, Tissue distribution of TRX f, TRX m, and FTR mRNA isolated from pea leaves (L), stems (St), roots (R), seeds (S), and flowers (F). Absolute mRNA levels of all genes were determined by semiquantitative RT-PCR. cDNA was standardized by reference to an actin standard. B, Western blotting analysis of different pea tissue extracts. PsTrxm, Antibodies against pea TRX m. PsTrxf, Antibodies against pea TRX f. Twenty micrograms of protein were loaded in each lane from tissue extracts of seeds (S), flowers (F), leaves (L), roots (R), and pods (P).
Table I.
Semiquantitative data for TRX f and TRX m expression in different Arabidopsis and pea tissues and organs, determined with four techniques
These data summarize the observations of transgenic plant lines and pea tissues. +++, High; ++, moderate; +, low; nd, not determined.
| Leaf
|
Root
|
Flower
|
Seed
|
Fruit
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| TRX f | TRX m | TRX f | TRX m | TRX f | TRX m | TRX f | TRX m | TRX f | TRX m | |
| GUS staining (Arabidopsis) | +++ | +++ | ++/+ | ++/+ | +++ | +++ | ++ | nd | ++ | ++ |
| ISH method (pea) | +++ | +++ | ++ | ++ | ++ | ++ | ++ | ++ | nd | nd |
| mRNA expression (pea) | ++ | +++ | + | + | ++ | +++ | ++ | +++ | nd | nd |
| Western blotting (pea) | +++ | +++ | + | ++ | ++ | ++ | ++ | ++ | +++ | ++ |
Western Blotting Identified TRX f and TRX m Protein in Root and Flower Tissues
Immunoblot analyses were performed with a pea anti-TRX m and anti-TRX f serum. Seeds, flowers, leaves, roots, and pod extracts from pea showed a clear band corresponding to TRX m recognized by the antibody, with a molecular mass of 14 kD (Fig. 8B). Similar results were obtained with anti-TRX f serum (Fig. 8B). Chloroplastic TRXs m and f were detected in all tissues analyzed (Table I).
Table I summarizes the sites of expression and level of expression observed with four different techniques that yield information on all steps of gene expression. The findings show clearly that the localization as well as the level of expression were similar in organs assayed.
DISCUSSION
Since the discovery of chloroplastic TRXs in nonphotosynthetic organs, significant efforts have been focused on the search for protein targets for all plant chloroplastic TRXs in an attempt to assign specific functions to each protein. To attain better insights into the potential role of chloroplastic TRXs, we first analyzed the spatial and temporal distribution of these proteins by inserting fusions of the GUS gene with the pea TRX f and TRX m promoters into Arabidopsis plants to trace the localization and pattern of gene expression of pea TRXs f and m. The results obtained were informative for new sites of action and are potentially useful in assigning different roles to chloroplastic TRXs.
During germination of PsTRXf1∷GUS transgenic plants, the pattern of GUS expression showed that TRX f is localized in areas of intensive cell division and confirmed previous studies that found TRX f mRNA in roots. These data suggest that pea TRX f might be involved in cell proliferation. We observed that, at later stages of growth, the intensity of GUS expression decreased at the beginning of senescence, indicating that chloroplastic TRX f is not related to the senescence process. In contrast, however, Laloi et al. (2004) found that AtTRX h5 was overexpressed in old leaves; these authors accordingly related this gene to senescence rather than to cell division. We have also detected TRX f in guard cells of stomata. Among the numerous proteins localized in guard cells, ADP-Glc pyrophosphorylase (AGPase), which is redox regulated, seems to be reduced by TRX f (Ballicora et al., 2000). Surprisingly, however, we detected GUS staining in stigmas, as well as in pollen grains in floral organs. Cytosolic TRXs have been isolated from pollen and were reported to take part in the self-incompatibility process in pollen-pistil recognition of Brassica oleracea (Bower et al., 1996; Cabrillac et al., 2001); however, the role of AtTRX h in pollen is still poorly understood in Arabidopsis, where self-incompatibility does not occur. Some authors have shown that the allergic response to grass pollen is linked to TRX h, which may thus represent a novel family of cross-reactive allergens that might contribute to the symptoms of baker's asthma (Weichel et al., 2006). In contrast to TRX h, however, no previous studies have found chloroplastic TRXs to be related with allergies. Despite the presence of Arabidopsis chloroplastic TRX y1 (Collin et al., 2004) and TRX m3 (Mestres-Ortega and Meyer, 1999) in flower buds, no previous studies have mentioned the existence of TRX f in reproductive organs.
Analysis of the TRX f promoter identified three cis-elements specific to root localization (ATATT) and at least five light-regulated motifs that are important for spatially regulated expression during normal plant development. Consistent with a regulatory role of these cis-acting sequences is our finding that GUS activity was not induced in transgenic plants with the truncated 149-bp promoter.
As with the TRX f promoter, in seedlings and 15-d-old Arabidopsis plants, the entire TRX m promoter directed strong GUS activity in regions of intense cell division, such as the cotyledons and leaf apical meristem. In fully developed transgenic plants, the intensity of GUS activity decreased as senescence progressed; however, GUS staining became stronger in the apical root meristem, at the ends of the siliques, and in flowers, pollen grains, and seeds. Like TRX f, the activity of TRX m may be directly related to the redox regulation of processes in tissues with high proportions of cells undergoing division. Serial deletions of 5′ fragments showed that a 259-bp segment of the TRX m proximal promoter (PsTRXm4∷GUS) was sufficient to confer a pattern of localization of GUS expression similar to that seen with PsTRXm1∷GUS. It therefore appears likely that the remaining 5′ flanking sequence of PsTRXm5∷GUS (97 bp) that harbors the putative TATA box and two light-dependent elements is largely repressed in Arabidopsis transformants.
In contrast to our findings with the PsTRXm1∷GUS fusion, experiments with plants carrying PsTRXm2∷GUS or PsTRXm3∷GUS (lacking the −1,874 and −1,521 and −1,874 and −503 sequences, respectively) led to staining of the hypocotyl and cotyledons, indicating the existence of regulatory cis-elements between nucleotides −1,874 and −503 that repressed expression in the hypocotyl. This finding reflected a tissue-dependent mechanism of control of gene expression. Deletion of this region may also have contributed to the high level of expression of PsTRXm2 and PsTRXm3∷GUS in comparison to PsTRXm1∷GUS. Taken together, these results indicate that the region between −1,874 and −503 bp harbors positive and negative regulatory elements that coordinate the tissue-specific regulation as well as the intensity of TRX m expression. Removing the sequence between −503 and −138 bp to obtain PsTRXm4∷GUS also led to expression of the TRX m promoter in the hypocotyl and cotyledons and to higher GUS activity. Although we had expected removal of light-regulatory motifs to decrease GUS expression when plants were subjected to light, serial deletions of 5′ fragments in the TRX m promoter led to an increase in the light-induced level of GUS mRNA, such that PsTRXm4∷GUS expression was more than 5-fold as high as PsTRXm1∷GUS expression, and PsTRXm2∷GUS expression was twice as high as GUS PsTRXm1∷GUS expression. Even more interesting, however, was the finding that the loss of GUS expression did not correlate with the transition to darkness. The loss of light regulation suggests that the fragment between −1,521 and −138 bp, which harbors eight light-related cis-elements, is required for the controlled increase and decrease in expression when plants are subjected to light-to-dark transition.
Nevertheless, it is also possible that chloroplastic TRXs are regulated not only by light, but also by other nutritional or hormonal factors that might influence their patterns of expression. These factors could explain why expression was not completely lost during darkness. Because chloroplastic TRXs regulate enzymes involved in carbon metabolism, it is likely that sugars are an essential factor that controls the transcription of the TRXs. Analysis of the TRX f and TRX m gene promoters fused to the GUS reporter gene revealed that environmental factors and tissue-specific control of TRX gene expression require complex integration of multiple cis-acting regulatory elements with different trans-acting proteins. However, accurate analyses of the promoters involved will be necessary to identify overrepresented motifs that might mediate organ-specific expression and the response to light and other stimuli. Currently, we are working on the research of expression pattern under different nutritional and light conditions.
Both TRX f and m are found in heterotrophic tissues and, like some of the cytosolic TRXs, chloroplastic TRX proteins might be involved in the redox control of cell proliferation during growth, as observed with the tobacco (Nicotiana tabacum) TRX h1 (Brugidou et al., 1993; Reichheld et al., 1999). No specific role has been established for this physiological process. However, in recent years, a number of new protein targets linked to regulatory disulfide protein TRXs have been identified, and these targets have helped to elucidate their function in chloroplasts (Motohashi et al., 2001, 2003; Balmer et al., 2003). Among them are the proteins involved in nitrogen metabolism, the C4 cycle, the translation process, fatty acid biosynthesis, and the 2-Cys peroxiredoxin stress-related protein. Proteins that might be related to the specific expression of chloroplastic TRX f and TRX m in areas of high cell division have been grouped together with proteins involved in folding and assembly (70-kD heat shock protein) and plastid division (FtsZ protein), and with proteins that play a role in DNA replication and transcription (ATP-dependent DNA helicase; Balmer et al., 2003).
Our findings in roots and flowers prompted a more detailed study of these tissues. We carried out in situ hybridization experiments that showed, as expected, that most TRX f and TRX m transcripts were expressed in the mesophyll cells of the leaves and around the vascular tissues. Hybridization experiments also confirmed the presence of TRX f and TRX m in roots, where TRXs were intensely expressed around the xylem and phloem. Because nutrients, water, ions, proteins, lipids, hormones, and other substances are transported through the xylem and phloem, it is difficult to identify a specific protein target in these tissues. Nevertheless, the role of chloroplastic TRXs is likely related to the complex transport system in vascular tissue, where these proteins may be involved in redox regulation of some membrane transporters. Support for this hypothesis is provided by the finding with proteomic methods that the plastidial ADP-Glc transporter (Brittle-1 protein) was a potential protein target of the spinach (Spinacia oleracea) TRX m (Balmer et al., 2006a).
TRX expression in developing pollen grains and embryos seems to correspond to a developmental requirement, as well as to nutritional status. Within the pollen grain, TRX m was detected as two distinct signals, probably situated in vegetative and generative cells. The pollen grain cells contain several organelles, endoplasmic reticulum, and plastids with starch used in pollen tube formation and storage substances such as lipids, proteins, and vitamins. The precise function of TRXs in floral organs is not yet clear, but it appears evident that chloroplastic TRXs are involved in redox regulation of some processes related to ovule and pollen grain maturation, fertilization, and embryo formation. The occurrence of both TRXs in the developing embryo is likely related to a role in the process of cell division during seed formation. In seeds, chloroplastic TRXs have been detected in an area of very active cell division, indicating an important function during germination. Montrichard and colleagues (2003) reported a differential pattern of expression of pea cytosolic TRX h3 and TRX h4 in early seedlings and seeds and suggested that these proteins were closely linked to germination. As found for other TRX h subtypes, chloroplastic TRXs might play a role in activating enzymes involved in storage protein mobilization during early seedling growth, as well as in carbohydrate metabolism.
To confirm these new sites of localization, we carried out immunological studies with antibodies against TRX f and TRX m. This approach detected protein in leaves, roots, pods, seeds, and flowers, and the amount of protein correlated with the level of mRNA expression in the same tissues (Table I). Recently, TRX m was detected with spinach TRX m antibodies in the endosperm amyloplast of wheat seeds (Balmer et al., 2006a). The question is thus whether the role of chloroplastic TRX expressed in reproductive organs and in roots is similar to that of the protein involved in regulating enzymes of the Calvin cycle in green tissue or in storage protein mobilization during seed germination (Kobrehel et al., 1992; Besse et al., 1996; Wong et al., 2002; Marx et al., 2003), or whether the TRX signal we observed is involved in self-incompatibility control in pollen-pistil recognition (Bower et al., 1996; Cabrillac et al., 2001), as has been observed for TRX h. Although our findings thus far in roots and flowers are insufficient to explain the specific role of TRXs in these parts of the plant, they nonetheless offer intriguing clues as to the possible functions of these proteins.
In relation to the role of plastid TRX in heterotrophic tissues during carbon metabolism, previous research reported low levels of FBPase and MDH transcripts and proteins in seeds and roots, but no enzyme activities were detected in these organs (Pagano et al., 2000). However, the fact that roots, pollen grains, and ovaries contain plastids and amyloplasts, which store starch and other important components, suggests that the role of TRX f and TRX m in roots and floral organs may lie in the redox regulation of enzymes involved in starch and carbohydrate metabolism. Additional support for the role of plastidial TRX in nonphotosynthetic tissues comes from the earlier finding that redox activation of AGPase in potato (Solanum tuberosum) tubers by chloroplastic TRX is part of a general mechanism for the regulation of starch synthesis in response to carbon status (Ballicora et al., 2000). Furthermore, Sparla and colleagues (2006) found transcripts of TRX-regulated β-amylase in leaves, roots, flowers, pollen, and seeds.
These findings suggest that TRX redox regulation is not a mechanism of regulation that occurs exclusively in photosynthetic tissues, but is rather a system common to every plastid in the plant (Tiessen et al., 2002, 2003; Geigenberger et al., 2005). In nonphotosynthetic organs, ferredoxin may not be reduced by light through the ferredoxin-TRX system, as is the case in chloroplasts, but in studies that used novel and improved methods, Balmer and colleagues (2006a) identified a complete ferredoxin-TRX system in amyloplasts from wheat starchy endosperm that involves ferredoxin, ferredoxin-NADP reductase, and FTR. As a mechanism of regulation, these authors suggested that light might be recognized as a thiol signal in the chloroplast, where it allows synthesis of sugar to proceed during photosynthesis. Balmer and colleagues further proposed that the synthesized sugar was then transported to sink organs, such as the seeds, and hydrolyzed by Glc-6-P dehydrogenase and 6-phosphogluconate dehydrogenase to generate NADPH, a thiol signal needed to reduce ferredoxin with ferredoxin-NADP reductase in the amyloplast (Balmer et al., 2006a). The reactions taking place in the amyloplast would thus be indirectly controlled by photosynthesis.
Because roots and floral organs also contain amyloplasts and starchy substances, we propose that they might also be equipped with a complete ferredoxin-TRX system. We detected the presence of pea FTR together with TRX f and TRX m mRNAs in leaves, stems, roots, flowers, and seeds, and suggest that in nonphotosynthetic tissues FTR is probably the enzyme that reduces TRXs. However, it will be necessary to identify other components of the ferredoxin-TRX system in roots and flowers before more can be said about the redox mechanisms through TRXs in the chloroplast of these organs. A search of Genevestigator microarray gene expression databases revealed similar patterns of expression of Arabidopsis chloroplastic TRXs in the tissues and organs assayed in our experiments (Zimmermann et al., 2004) and thus supported the new localizations we found for these proteins.
In relation with plastidial TRX functions, the identification of potential protein targets in wheat endosperm amyloplasts has helped to determine the possible roles of the disulfide protein in heterotrophic organs. These proteins are classified according to the process in which they are involved (Balmer et al., 2006b). Among 289 proteins, we found proteins involved in starch and carbohydrate metabolism, lipid and amino acid biosynthesis, amino acid metabolism, and protein assembly and folding. In addition to AGPase, other protein targets have been biochemically linked to TRX (e.g. acetyl-CoA carboxylase [Sasaki et al., 1997], cyclophilin [Motohashi et al., 2003], peroxiredoxin BAS1 [König et al., 2002], protein disulfide isomerase [Danon and Mayfield, 1994; Kim and Mayfield, 2002], and thiosulfate sulfurtransferase [Ray et al., 2000]). In nonphotosynthetic tissues, chloroplastic TRXs appear to be related with carbohydrate and lipid metabolism and amino acid synthesis, DNA replication and translation, protein folding, and protein storage mobilization, rather than with photosynthesis.
In conclusion, this work provides strong evidence that TRX f and TRX m proteins are localized in roots, vascular tissues, seeds, and pollen grains and embryos in reproductive organs, and seems to participate in all stages of plant development. In addition, our results strongly suggest that chloroplastic TRX f and TRX m are involved in cell division and plant reproduction. By analyzing TRX f and TRX m promoters we were able to localize several functional domains necessary for light-dependent expression and negative regulation of tissue-specific expression of TRX m in the hypocotyls of Arabidopsis plants. Future work will concentrate on (1) analyzing the specific function of these proteins in nonphotosynthetic tissues to elucidate their role in different organs; (2) identifying specific protein targets for each plastidial TRX in plant parts that have yet to be studied, especially in heterotrophic tissues and flowers; and (3) assigning definite physiological functions to these proteins in different stages of development and growth of the plant. It is hoped that our results will contribute to the understanding of the physiological roles of other TRXs in different parts of the plant.
MATERIALS AND METHODS
Plant Material
Pea (Pisum sativum var. Lincoln) leaves, stems, roots, flowers, and seeds were used in protein, DNA, and RNA extraction and for in situ hybridization experiments. Plants were grown in a green cabinet for 15 d, first in vermiculite, and then in compost until flowering.
Arabidopsis (Arabidopsis thaliana) ecotype Columbia and transgenic seeds were germinated in compost and vermiculite (compost:vermiculite [2:1]) under controlled environmental conditions: 140 μmol photons m−2 s−1 photosynthetically active radiation; 23°C temperature; 16-h-light/8-h-dark photoperiod, and daily watering with a nutrient solution for Arabidopsis. For in vitro germination, we used Murashige and Skoog medium described by Murashige and Skoog (1962) supplemented with vitamins and the appropriate antibiotic (kanamycin) for the transgenic lines.
Isolation of Promoter DNA
The promoters of the TRX f and TRX m genes were isolated by PCR walking according to the protocol described by Devic et al. (1997). Approximately 2.5 μg of pea genomic DNA were digested overnight with 50 units of each enzyme at a final volume of 100 μL. Seven restriction enzymes (EcoRV, HindIII, ScaI, PvuII, DraI, HpaI, and StuI) to create blunt-ended fragments were used independently for library construction. The adaptor was ligated to genomic DNA overnight at 16°C in the presence of 10 units of T4 DNA ligase (Promega) in a final reaction volume of 20 μL. Adaptor primers were AP1, 5′-GGATCCTAATACGACTCACTATAGGGC-3′; and AP2, 5′-CTATAGGGCTCGAGCGGC-3′. For nested PCR reactions, Long Expand Taq polymerase mix (Boehringer) was used in combination with oligonucleotides specific for TRX f and TRX m. For the TRX f promoter, we used F1, 5′-ACCTCTGTAGACCAACCCTCTTCG-3′; and F2, 5′-GGTGCCGATCCATTTAGGGGAGGTG-3′. For the TRX m promoter, we used M1, 5′-GGAAGACAATGAGGTCTTGGTGTGG-3′; and M2, 5′-ACTCTTAAACAAGCTCTCAAGGGCC-3′.
The primary PCR cycling parameters were seven cycles at 94°C for 1 min and 72°C for 4 min, then 32 cycles at 94°C for 1 min and 65°C for 4 min, and the oligos used were AP1 and F1 for TRX f and AP1 and M1 for TRX m. For the second PCR, the program was five cycles at 94°C for 1 min and 72°C for 4 min, then 25 cycles at 94°C for 1 min and 65°C for 4 min, and the oligos were AP2 and F2 for TRX f and AP2 and M2 for TRX m. The fragments obtained were subcloned into the plasmid pGEM-T Easy vector (Promega) for sequencing.
Database-Assisted Promoter Analysis
Putative cis-regulatory sequences were sought with bioinformatics approaches using two databases that identified transcription factor binding sites or cis-acting sequences in plant promoters: PLACE (http://www.dna.affrc.go.jp/htdocs/PLACE; Higo et al., 1999) and Plant CARE (http://sphinx.rug.ac.be:8080/PlanCARE/index.html; Rombauts et al., 1999). We also analyzed the promoter regions of the two TRXs f and four TRXs m of Arabidopsis located in the TAIR database. To analyze the Genevestigator databases for microarray gene expression, we used the tools at https://www.genevestigator.ethz.ch (Zimmermann et al., 2004).
RNA Extraction and Transcription Start Sites
The transcription start point was determined by reverse transcriptase-mediated RACE-PCR with a FirstChoice RLM-RACE kit (Ambion) according to the manufacturer's instructions. Total RNA (12 μg) extracted from pea leaves was isolated with TRIzol (BD) and used for RT. 5′-RACE-PCR was done in two steps with nested gene-specific inner and outer primers (F1 and F2 for TRX f; M1 and M2 for TRX m) and nested primers from the 5′-RACE adaptor provided in the kit. Annealing temperatures for the reactions were 60°C (outer) and 65°C (inner). For sequencing, the PCR products were cloned into pGEM-T Easy vector (Promega).
RT and Semiquantitative PCR Analysis
First-strand cDNA was synthesized from 2.5 μg of total RNA using SuperScript III reverse transcriptase (Invitrogen) and oligo(dT)12-15 primer. The reaction was incubated at 50°C for 1 h. To analyze expression, we used the pea actin gene as a housekeeping gene. The PCR reaction conditions were 2 min at 94°C, 30 cycles at 92°C for 30 s, 60°C for 30 s, 72°C for 30 s, and 72°C for 5 min. The specific oligonucleotides were 5′-TGATAAAACCGTCGTCCTCGAT-3′ for Trxf-F, 5′-ATTTCCTCATCTTCCCCTCAGC-3′ for Trxf-R, 5′-GTTTCACTTCGCTGGTGTTGC-3′ for Trxm-F, 5′-CTTTCTCAGACAGAGTAGCC-3′ for Trxm-R, 5′-CAAGCTTCAACCTTCGCCGTC-3′ for FTR-F, 5′-TGGCACTCCTTTCTCTCTCTC-3′ for FRT-R, 5′-AATGGTGAAGGCTGGATTTG-3′ for Act-F, and 5′-AGCAAGATCCAAACGAAGGA-3′ for Act-R.
GUS Reporter Gene Construction and Expression in Transgenic Arabidopsis Plants
Full-length fragments were amplified by PCR between positions ATG and −444 for TRX f and between ATG and −1,874 for TRX m with oligonucleotides bearing appropriate restriction sites (SalI and BamHI), and subcloned into the pBI101 vector to produce the constructions PsTRXf1 and PsTRXm1. Genomic fragments of 444 bp (TRX f) and 1,973 bp (TRX m) were used to construct a series of 5′ deletions of the two chloroplastic TRX promoters fused to the GUS reporter gene. One 5′ deletion fragment for the TRX f promoter extending from positions ATG to −126 (PsTRXf2) and four 5′ deletion fragments for the TRX m promoter from positions ATG to −1,521 (PsTRXm2), ATG to −503 (PsTRXm3), ATG to −138 (PsTRXm4), and ATG to +23 (PsTRXm5) were also produced by DNA amplification with Long Expand Taq polymerase mix. To obtain the deleted promoter fragments by PCR, we used the following primers (Fig. 1): prTrxf1, 5′-GTGTCGACGTAAACAAATAAACCACAT-3′; prTrxf2, 5′-GTGGATCCCATTGATGTGGGGAAGA-3′; prTrxf3, 5′-GTGTCGACCCATACACAGACTTCAA-3′; prTrxm1, 5′-GTGTCGACCTCCTTGGTGCCTCTCAA-3′; prTrxm2, 5′-GTGGATCCCATGTTATTTTCTGTGTAT-3′; prTrxm3, 5′-GTGTCGACGTTCATACTTTTTCCCT-3′; prTrxm4, 5′-GTGTCGACGTGTTTAGGGTACATTC-3′; prTrxm5, 5′-GTGTCGACGTGTGGAGAAGTACTT-3′; and prTrxm6, 5′-GTGTCGACCCGCCACAACACTAG-3′. Restriction sites are shown in bold (SalI) and italics (BamHI).
PCR products were ligated in-frame to the GUS reporter gene in the binary vector pBI101 using the restriction sites SalI and BamHI. Seven constructs were generated: PsTRXf1∷GUS (444 bp), PsTRXf2∷GUS (126 bp), PsTRXm1∷GUS (1973 bp), PsTRXm2∷GUS (1642 bp), PsTRXm3∷GUS (624 bp), PsTRXm4∷GUS (259 bp), and PsTRXm5∷GUS (89 bp; Fig. 1). The promoter region of all constructs was confirmed by sequencing. Plasmids were introduced into Agrobacterium tumefaciens C58C1Rif and used to transform Arabidopsis plants by the floral-dip method (Bechtold et al., 1993). Seeds from infiltrated plants were sterilized for 1 min with 70% ethanol and 5 min with a mixture of 5% sodium hypochlorite and 2.5% Tween 20, and then rinsed three times with sterile water and germinated on Murashige and Skoog medium with the addition of 50 μg mL−1 kanamycin. Seedlings were grown in a light cabinet at 23°C and a 16-h-light/8-h-dark photoperiod, and the kanamycin-resistant plantlets were transplanted individually to sterile compost. Primary transformants were selfed and the homozygous seeds from the second or third generations (T2 or T3) were used for later experiments after cultivation under similar conditions. Arabidopsis plants carrying the pBI101 vector without promoter were used as a negative control for GUS expression. For the GUS assay, plants were grown in Murashige and Skoog medium supplemented with Suc, agar, and kanamycin in a light cabinet or in sterile compost in a greenhouse under a 16-h-light/8-h-dark regime. Samples were taken either from in vitro experiments or from soil every 4 h during the day and every 8 h during the night.
GUS Expression Analysis
Histochemical staining for GUS in transgenic Arabidopsis plants was performed according to standard procedures with minor modifications (Jefferson et al., 1987). Histochemical in situ localization of GUS expression was performed on five independent transgenic lines of the PsTRXf1∷GUS and PsTRXm1∷GUS constructs, and on two independent lines of each of the deletions. One kanamycin-resistant plant for the pBI101 binary vector was used as a control.
Localization of TRX f and TRX m Expression
Histochemical staining for GUS was performed at 37°C by incubating the material in a solution containing 1 mm 5-bromo-4-chloro-3-indolyl β-d-glucuronide as a substrate, 50 mm potassium phosphate, pH 7, 0.5 mm potassium ferrocyanide, 0.5 mm potassium ferricyanide, 0.1% (v/v) Tween 20, and 0.1% (v/v) Triton X-100. After staining, plants were incubated in 70% (v/v) ethanol at 37°C overnight. Plant materials used were either early seedlings grown on plates or different growth stages of the rosette and flowers of 4- to 5-week-old plantlets grown in soil. Pigments were extracted from stained tissues with ethanol:acetic acid (1:1). After extensive washing, the clarified tissues were stored in 50% (v/v) glycerol until photodocumentation. Micrographs were taken with an Olympus AX-70 microscope. Stained leaves were embedded in Unicryl (British BioCell International); semithin sections were obtained with a Leica RM2164 microtome. Sections were counterstained with toluidine blue or safranine dyes and observed with a Zeiss Axioplan microscope equipped with a CCD and micrographs were taken with a Canon PowerShot S40 camera.
In Situ Hybridization
In situ hybridization was performed according to earlier methods (Ferrandiz et al., 2000). The plasmids pTrxf and pTrxm, containing the open reading frame of TRX f and TRX m, respectively, were used to generate sense and antisense probes for in situ hybridization. Sense and antisense probes were labeled with digoxigenin-11-UTP (Roche Molecular Biochemical) according to the manufacturer's instructions. For the sense probe, pTrxf and pTrxm were linearized with SacII (KspI); for the antisense probes, pTrxf was linearized with SalI and pTrxm was linearized with SacI. The linearized probes were transcribed with either T7 RNA polymerase (TRX f sense and TRX m antisense) or SP6 RNA polymerase (TRX f antisense and TRX m sense). Tissue samples were fixed in FAE solution (ethanol:acetic acid:formaldehyde:water at 50:5:3.7:41.3 [v/v/v/v]), dehydrated, embedded in paraffin, and sectioned at 10 μm. Sections were hybridized with the different probes overnight at 50°C and incubation with the antibody and color detection were done according to the manufacturer's instructions (Roche). Micrographs were taken with an Olympus BX51 microscope and an Olympus DP50 camera.
Protein Extraction and Western Blotting
Flowers, seeds, leaves, roots, and pods were homogenized with 25 mm Tris-HCl, pH 7.5, and 5 mm MgCl2 (1:3 [w/v]). After 20 min of centrifugation at 12,000g, the supernatant was removed and the protein concentration determined with the Bradford assay (Bio-Rad), with bovine serum albumin as the standard (Bradford, 1976). Electrophoresis in polyacrylamide gel was performed according to Laemmli (1970), proteins were transferred to a nitrocellulose membrane, and western blotting was performed as in Towbin et al. (1979), using polyclonal antibodies against pea-leaf chloroplastic TRX m diluted 1:1,000 or TRX f diluted 1:1,000 (v/v). Bound antibody was visualized with enhanced chemiluminescence (ECL system; Amersham Biosciences).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers PsTRXf (EF483938) and PsTRXm (EF483939).
Acknowledgments
We thank C. Ferrandiz for help with developing the in situ hybridization technique, T. Moreno for technical assistance, and K. Shashok for helpful editorial feedback on the manuscript.
This work was supported by the Ministerio de Educación y Ciencia (Spain; grant nos. BFI2002–00401 and BIO2005–00157) and the Junta de Andalucía (Spain; grant no. CVI–154). J.d.D.B.-L. was supported by a predoctoral fellowship from the Junta de Andalucía and A.S.R. was supported by a postdoctoral I3P fellowship from the Consejo Superior de Investigaciones Científicas (Spain).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Mariam Sahrawy (mariam.sahrawy@eez.csic.es).
Open Access articles can be viewed online without a subscription.
References
- Arner ES, Holmgren A (2000) Physiological functions of thioredoxin and thioredoxin reductase. Eur J Biochem 267 6102–6109 [DOI] [PubMed] [Google Scholar]
- Ballicora MA, Frueauf JB, Fu Y, Schürmann P, Preiss J (2000) Activation of the potato tuber ADP-glucose pyrophosphorylase by thioredoxin. J Biol Chem 275 1315–1320 [DOI] [PubMed] [Google Scholar]
- Balmer Y, Koller A, del Val G, Manieri W, Schürmann P, Buchanan BB (2003) Proteomics gives insight into the regulatory function of chloroplast thioredoxins. Proc Natl Acad Sci USA 100 370–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balmer Y, Vensel WH, Cai N, Manieri W, Schürmann P, Hurkman WJ, Buchanan BB (2006. a) A complete ferredoxin/thioredoxin system regulates fundamental processes in amyloplasts. Proc Natl Acad Sci USA 103 2988–2993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balmer Y, Vensel WH, DuPont FM, Buchanan BB, Hurkman WJ (2006. b) Proteome of amyloplasts isolated from developing wheat endosperm presents evidence of broad metabolic capability. J Exp Bot 57 1591–1602 [DOI] [PubMed] [Google Scholar]
- Balmer Y, Vensel WH, Tanaka CK, Hurkman WJ, Gelhaye E, Rouhier N, Jacquot JP, Manieri W, Schürmann P, Droux M, et al (2004) Thioredoxin links redox to the regulation of fundamental processes of plant mitochondria. Proc Natl Acad Sci USA 101 2642–2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassham JA, Krause GH (1969) Free energy changes and metabolic regulation in steady-state photosynthetic carbon reduction. Biochim Biophys Acta 189 207–221 [DOI] [PubMed] [Google Scholar]
- Bechtold N, Ellis J, Pelletier G (1993) In planta Agrobacterium-mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C R Acad Sci Ser III Sci Vie 316 1194–1199 [Google Scholar]
- Besse I, Wong JH, Kobrehel K, Buchanan BB (1996) Thiocalsin: a thioredoxin-linked, substrate-specific protease dependent on calcium. Proc Natl Acad Sci USA 93 3169–3175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bower MS, Matias DD, Fernandes-Carvalho E, Mazzurco M, Gu T, Rothstein SJ, Goring DR (1996) Two members of the thioredoxin-h family interact with the kinase domain of a Brassica S locus receptor kinase. Plant Cell 8 1641–1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72 248–254 [DOI] [PubMed] [Google Scholar]
- Brugidou C, Marty I, Chartier Y, Meyer Y (1993) The Nicotiana tabacum genome encodes two cytoplasmic thioredoxin genes which are differently expressed. Mol Gen Genet 238 285–293 [DOI] [PubMed] [Google Scholar]
- Cabrillac D, Cock JM, Dumas C, Gaude T (2001) The S-locus receptor kinase is inhibited by thioredoxins and activated by pollen coat proteins. Nature 410 220–223 [DOI] [PubMed] [Google Scholar]
- Carrasco JL, Chueca A, Sahrawy M, Hermoso R, Lázaro JJ, López Gorgé J (1992) Role of light in the in vivo and in vitro synthesis of spinach thioredoxin f. Physiol Plant 84 236–242 [Google Scholar]
- Collin V, Issakidis-Bourget E, Marchand C, Hirasawa M, Lancelin JM, Knaff DB, Miginiac-Maslow M (2003) The Arabidopsis plastidial thioredoxins: new functions and new insights into specificity. J Biol Chem 278 23747–23752 [DOI] [PubMed] [Google Scholar]
- Collin V, Lamkemeyer P, Miginiac-Maslow M, Hirasawa M, Knaff DB, Dietz KJ, Issakidis-Bourguet E (2004) Characterization of plastidial thioredoxins from Arabidopsis belonging to the new y-type1. Plant Physiol 136 4088–4095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford NA, Yee BC, Nishizawa AN, Buchanan BB (1979) Occurrence of cytoplasmic f- and m-type thioredoxins in leaves. FEBS Lett 104 141–145 [Google Scholar]
- Danon A, Mayfield SP (1994) Light-regulated translation of chloroplast messenger RNAs through redox potential. Science 266 1717–1719 [DOI] [PubMed] [Google Scholar]
- Devic M, Albert S, Delseny M, Roscoe TJ (1997) Efficient PCR walking on plant genomic DNA. Plant Physiol Biochem 35 331–339 [Google Scholar]
- Elmayan T, Tepfer M (1995) Evaluation in tobacco of the organ specificity and strength of the rol D promoter, domain A of the 35S promoter and the 35S2 promoter. Transgenic Res 4 388–396 [DOI] [PubMed] [Google Scholar]
- Ferrandiz C, Gu Q, Martienssen R, Yanofsky MF (2000) Redundant regulation of meristem identity and plant architecture by FRUITFULL, APETALA1 and CAULIFLOWER. Development 127 725–734 [DOI] [PubMed] [Google Scholar]
- Geck MK, Larimer FW, Hartman FC (1996) Identification of residues of spinach thioredoxin f that influence interactions with target enzymes. J Biol Chem 271 24736–24740 [DOI] [PubMed] [Google Scholar]
- Geigenberger P, Kolbe A, Tiessen A (2005) Redox regulation of carbon storage and partitioning in response to light and sugars. J Exp Bot 56 1469–1479 [DOI] [PubMed] [Google Scholar]
- Gilmartin PM, Sarokin L, Memelink J, Chua NH (1990) Molecular light switches for plant genes. Plant Cell 2 369–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higo K, Ugawa Y, Iwamoto M, Korenaga T (1999) Plant cis-acting regulatory DNA elements (PLACE) database. Nucleic Acids Res 27 297–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges M, Miginiac-Maslow M, Decottignies P, Jacquot JP, Stein M, Lepiniec L, Crétin C, Gadal P (1994) Purification and characterization of pea thioredoxin f expressed in Escherichia coli. Plant Mol Biol 26 225–234 [DOI] [PubMed] [Google Scholar]
- Holmgren A (1985) Thioredoxin. Annu Rev Biochem 54 237–271 [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW (1987) Gus fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Mayfield SP (2002) The active site of the thioredoxin-like domain of chloroplast protein disulfide isomerase, RB60, catalyzes the redox-regulated binding of chloroplast poly(A)-binding protein, RB47, to the 5′ untranslated region of psbA mRNA. Plant Cell Physiol 43 1238–1243 [DOI] [PubMed] [Google Scholar]
- Kobrehel K, Wong JH, Balogh A, Kiss F, Yee BC, Buchanan BB (1992) Specific reduction of wheat storage proteins by thioredoxin h. Plant Physiol 99 919–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- König J, Baier M, Horling F, Kahmann U, Harris G, Schürmann P, Dietz KJ (2002) The plant-specific function of 2-Cys peroxiredoxin-mediated detoxification of peroxides in the redox-hierarchy of photosynthetic electron flux. Proc Natl Acad Sci USA 99 5738–5743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227 680–685 [DOI] [PubMed] [Google Scholar]
- Laloi C, Mestres-Ortega D, Marco Y, Meyer Y, Reichheld JP (2004) The Arabidopsis cytosolic thioredoxin h5 gene induction by oxidative stress and its W-box-mediated response to pathogen elicitor. Plant Physiol 134 1006–1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam E, Chua NH (1989) ASF-2: a factor that binds to the cauliflower mosaic virus 35S promoter and a conserved GATA motif in Cab promoters. Plant Cell 1 1147–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Jaramillo J, Chueca A, Jacquot JP, Hermoso R, Lázaro JJ, Sahrawy M, López-Gorgé J (1997) High-yield expression of pea thioredoxin m and assessment of its efficiency in chloroplast fructose-1,6-bisphosphatase activation. Plant Physiol 114 1169–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marx C, Wong JH, Buchanan BB (2003) Thioredoxin and germinating barley: targets and protein redox changes. Planta 216 454–460 [DOI] [PubMed] [Google Scholar]
- Mestres-Ortega D, Meyer Y (1999) The Arabidopsis thaliana genome encodes at least four thioredoxins m and a new prokaryotic-like thioredoxin. Gene 240 307–316 [DOI] [PubMed] [Google Scholar]
- Meyer Y, Reichheld JP, Vignols F (2005) Thioredoxins in Arabidopsis and other plants. Photosynth Res 86 419–433 [DOI] [PubMed] [Google Scholar]
- Meyer Y, Vignols F, Reichheld JP (2002) Classification of plant thioredoxins by sequence similarity and intron position. Methods Enzymol 347 394–402 [DOI] [PubMed] [Google Scholar]
- Montrichard F, Renard M, Alkhalfioui F, Duval FD, Macherel D (2003) Identification and differential expression of two thioredoxin h isoforms in germinating seeds from pea. Plant Physiol 132 1707–1715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motohashi K, Kondoh A, Stumpp MT, Hisabori T (2001) Comprehensive survey of proteins targeted by chloroplast thioredoxin. Proc Natl Acad Sci USA 98 11224–11229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motohashi K, Koyama F, Nakanishi Y, Ueoka-Nakanishi H, Hisabori T (2003) Chloroplast cyclophilin is a target protein of thioredoxin. Thiol modulation of the peptidyl-prolyl cis-trans isomerase activity. J Biol Chem 278 31848–31852 [DOI] [PubMed] [Google Scholar]
- Mouaheb N, Thomas D, Verdoucq L, Monfort P, Meyer Y (1998) In vivo functional discrimination between plant thioredoxins by heterologous expression in the yeast Saccharomyces cerevisiae. Proc Natl Acad Sci USA 95 3312–3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15 473–497 [Google Scholar]
- Pagano EA, Chueca A, López-Gorge J (2000) Expression of thioredoxins f and m and of their targets fructose-1,6-bisphosphatase and NADP-malate dehydrogenase, in pea plants grown under normal and light/temperature stress conditions. J Exp Bot 51 1299–1307 [PubMed] [Google Scholar]
- Ray WK, Zeng G, Potters MB, Mansuri AM, Larson TJ (2000) Characterization of a 12-kilodalton rhodanese encoded by glpE of Escherichia coli and its interaction with thioredoxin. J Bacteriol 182 2277–2284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichheld JP, Vernoux T, Lardon F, Van Montagu M, Inzé D (1999) Specific checkpoints regulate plant cell cycle progression in response to oxidative stress. Plant J 17 647–656 [Google Scholar]
- Rombauts S, Déhais P, Van Montagu M, Rouzé P (1999) Plant CARE, a plant cis-acting regulatory element database. Nucleic Acids Res 27 295–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahrawy M, Hecht V, López-Jaramillo J, Chueca A, Chartier Y, Meyer Y (1996) Intron position as an evolutionary marker of thioredoxins and thioredoxin domains. J Mol Evol 42 422–431 [DOI] [PubMed] [Google Scholar]
- Sasaki Y, Kozaki A, Hatano M (1997) Link between light and fatty acid synthesis: thioredoxin-linked reductive activation of plastidic acetyl-CoA carboxylase. Proc Natl Acad Sci USA 94 11096–11101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schürmann P, Jacquot JP (2000) Plant thioredoxin systems revisited. Annu Rev Plant Physiol Plant Mol Biol 51 371–400 [DOI] [PubMed] [Google Scholar]
- Shirsat A, Wilford N, Croy R, Boulter D (1989) Sequences responsible for the tissue specific promoter activity of a pea legumin gene in tobacco. Mol Gen Genet 215 326–331 [DOI] [PubMed] [Google Scholar]
- Sparla F, Costa A, Lo Schiavo F, Pupillo P, Trost P (2006) Redox regulation of a novel plastid-targeted β-amylase of Arabidopsis. Plant Physiol 141 840–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiessen A, Hendriks JH, Stit M, Branscheid A, Gibon Y, Farré EM, Geigenberger P (2002) Starch synthesis in potato tubers is regulated by post-translational redox-modification of ADP-glucose pyrophosphorylase: a novel regulatory mechanism linking starch synthesis to the sucrose supply. Plant Cell 14 2191–2213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiessen A, Prescha K, Bransheid A, Palacios N, McKibbin R, Halford NG, Geigenberger P (2003) Evidence that SNF1-related kinase and hexokinase are involved in separate sugar-signalling pathways modulating post-translational redox activation of ADP-glucose pyrophosphorylase in potato tubers. Plant J 35 490–500 [DOI] [PubMed] [Google Scholar]
- Towbin H, Staehelin T, Gordon J (1979) Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA 76 4350–4354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traverso JA, Vignols F, Cazalis R, Pulido A, Sahrawy M, Cejudo FJ, Meyer Y, Chueca A (2007) PsTRXh1 and PsTRXh2 are both pea h-type thioredoxins with antagonistic behavior in redox imbalances. Plant Physiol 143 300–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weichel M, Glaser AG, Ballmer-Weber BK, Schmid-Grendelmeier P, Crameri R (2006) Wheat and maize thioredoxins: a novel cross-reactive cereal allergen family related to baker's asthma. J Allergy Clin Immunol 117 676–681 [DOI] [PubMed] [Google Scholar]
- Wong JH, Cai N, Balmer Y, Tanaka CK, Vensel WH, Hurkman WJ, Buchanan BB (2004) Thioredoxin targets of developing wheat seeds identified by complementary proteomic approaches. Phytochemistry 65 1629–1640 [DOI] [PubMed] [Google Scholar]
- Wong JH, Kim YB, Ren PH, Cai N, Cho MJ, Hedden P, Lemaux PG, Buchanan BB (2002) Transgenic barley grain overexpressing thioredoxin shows evidence that the starchy endosperm communicates with the embryo and the aleurone. Proc Natl Acad Sci USA 99 16325–16330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagisawa S (2000) Dof1 and Dof2 transcription factors are associated with expression of multiple genes involved in carbon metabolism in maize. Plant J 21 281–288 [DOI] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W (2004) GENEVESTIGATOR. Arabidopsis Microarray Database and Analysis Toolbox. Plant Physiol 136 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]