Abstract
The initiation of SV40 (simian virus 40) DNA replication requires the co-operative interactions between the viral Tag (large T-antigen), RPA (replication protein A) and Pol (DNA polymerase α-primase) on the template DNA. Binding interfaces mapped on these enzymes and expressed as peptides competed with the mutual interactions of the native proteins. Prevention of the genuine interactions was accomplished only prior to the primer synthesis step and blocked the assembly of a productive initiation complex. Once the complex was engaged in the synthesis of an RNA primer and its extension, the interfering effects of the peptides ceased, suggesting a stable association of the replication factors during the initiation phase. Specific antibodies were still able to disrupt preformed interactions and inhibited primer synthesis and extension activities, underlining the crucial role of specific protein–protein contacts during the entire initiation process.
Keywords: DNA polymerase α-primase (Pol), large T-antigen, protein–protein interaction, replication protein A, simian virus 40 DNA replication (SV40 DNA replication)
Abbreviations: DBD-A, DNA-binding domain A; MBP, maltose-binding protein; OBD, origin-DNA-binding domain; Pol, DNA polymerase α-primase; RPA, replication protein A; H6-RPA, His6-tagged RPA; ssDNA, single-stranded DNA; SV40, simian virus 40; Tag, large T-antigen; TBE, Tris/borate/EDTA; Topo, topoisomerase I
INTRODUCTION
SV40 (simian virus 40) provides an effective model system to study eukaryotic DNA replication [1]. One viral protein, Tag (large T-antigen), orchestrates the entire replication of the viral mini-chromosome in primate cellular extracts [2]. Tag assembles as a double hexamer on its cognate origin sequences, distorts and bidirectionally unwinds the duplex DNA in an ATP-dependent manner and recruits other cellular replication proteins to assemble the replication fork. For the initiation reaction, three cellular proteins are involved, in addition to Tag. Topo (topoisomerase I) releases torsional stress induced by the helicase activity of Tag ahead of and behind the replication fork. The heterotrimeric RPA (replication protein A) is required to stabilize emerging ssDNA (single-stranded DNA) regions and in conjunction with Tag enables Pol (DNA polymerase α-primase) to synthesize and extend primers [3]. For subsequent steps, the host replication machinery provides all other necessary factors [4].
The co-ordination of the individual steps during the replication reaction relies on the modular design of replication factors and their organization into structural and functional domains [5]. Specific interactions have been attributed to certain subunits or domains of Tag, RPA, Pol and Topo, and the contact sites between these key players are crucial for the initiation process [6–11]. Tag amino acids within the OBD (origin-DNA-binding domain) have been identified to interact with sequences within the DBD-A (DNA-binding domain A) of the largest p70 subunit of RPA. The same region of p70 and stretches of its N-terminal domain contact the largest p180 subunit of Pol, which in turn binds the N-terminal and helicase domains of Tag via a region situated N-terminally of the catalytic polymerase domain. Some of the activities of the single replication factors correlate well with the physical interactions that they undergo [6,11–13]. However, the precise timing of these interactions, and the steps when they are required, are only starting to emerge [10,14].
In the present study, we have employed peptides that interfered with specific individual associations between Tag, RPA and Pol. Mutual interactions of these factors first became important during the assembly of a metastable pre-initiation complex. The transition to the initiation phase was accompanied by a tightening of these interactions within the initiation complex (primosome).
MATERIALS AND METHODS
Cloning of fusion peptides
Sequences for RPA, Tag and Pol were amplified from cloned genes using primer pairs R70.1N and R70.173C for peptide R1-173, R70.174N and R70.250C for peptide R174-250 (both RPA), T96.164N and T96.249C for peptide T164–249 (Tag), and P180.195N and P180.313C for peptide P195-313 (Pol). The numbers preceding ‘N’ and ‘C’ indicate the first N- and last C-terminally expressed amino acids of the resulting amplified sequences. The primers had the following sequences, with restriction sites used for cloning underlined: R70.1N, 5′-TTGGATCCCATATGGTCGGCCAGCTGAGCGA-3′; R70.174N, 5′-TTGGATCCTCACACACTTCTGGGGGAA-3′; R70.173C, 5′-TTGAATTCTCACAGGCTGGGACCTGCAG-3′; R70.250C, 5′-TTGAATTCTCACACTTCAATAAGAGGAAAGAACT-3′; T96.164N, 5′-TTGGATCCCATATGAAGGAAAAAGCTGCACT-3′; T96.249C, 5′-TTGGATCCTCATGGCAAACTTTCCTCAA-3′; P180.195N, 5′-TTGAATCCCATATGTCTGTGCACACCGCCAC-3′ and P180.313C, 5′-TTGGATCCTCAACCTTCTTGATCAATGT-3′.
The EcoRI/BamHI fragments were cloned into pBluescript KS II+ (Stratagene) to yield plasmids pBS(N)/RPA70(1-173), pBS(N)/RPA70(174-250), pBS(N)/TAG(164-249) and pBS(N)/POL(195-313) respectively and were confirmed by sequencing. The BamHI/SalI fragments were then subcloned into pMAL-c2 (New England Biolabs) to yield plasmids pMAL/RPA70.1-173, pMAL/RPA70.174-259, pMAL/TAG96.164-249 and pMAL/POL180.195-313.
Purification of replication proteins and MBP (maltose-binding protein) fusion peptides
SV40 Tag and the human Pol were expressed in Sf9 cells infected with recombinant baculoviruses and purified as described in [15,16]. His6-tagged Topo expressed in baculovirus was purified on TALON™-resin (Clontech) and provided by Kent Søe (Clinical Cell Biology, Veje Hospital, Veje, Denmark) [17]. Bacterially expressed His6-tagged RPA (H6-RPA) was purified as outlined for RPA [18]. MBP-fusion peptides were expressed and purified on amylose resin as described in [19].
Protein–protein interactions
For co-precipitation assays [20], 50 pmol of specific monoclonal antibodies (0.15 μg/pmol), PAb 101 for Tag [21], SJK237-87 for Pol [22] and anti-His tag (Qiagen) for H6-RPA were coupled with 20 μl of 50% (v/v) Protein G–agarose beads in 100 μl of binding buffer (50 mM Hepes/KOH, pH 7.9, 100 mM KCl, 7 mM MgCl2, 0.25% inositol, 0.25 mM EDTA and 0.05% Nonidet P40) for 1 h at 4 °C. Alternatively, 50 μl of polyclonal anti-RPA serum was used. The beads were then washed four times with 1 ml of wash buffer (30 mM Hepes/KOH, pH 7.9, 100 mM KCl and 7 mM MgCl2). Then, 25 pmol of Tag (0.096 μg/pmol), Pol (3.54 μg/pmol) or RPA (0.116 μg/pmol) was added to the corresponding beads in a 100 μl reaction volume in binding buffer. Alternatively, MBP-fusion peptides served as the bait and were coupled with amylose resin at a concentration of 1.25 pmol/μl. After 1 h at 4 °C to immobilize the bait protein, the beads were washed four times with 1 ml of wash buffer. The beads were then incubated with 25 pmol of the target protein in the absence or presence of a 25–100 molar excess of peptides over the target protein or 50 pmol of antibodies in 100 μl of reaction buffer containing 2% (w/v) BSA for 2 h at 4 °C. Alternatively, antibodies were added after the target protein was allowed to bind to determine their disruption capability on preformed protein–protein interactions. In these cases, incubation continued for another 1 h. After washing four times as before, the beads were boiled in 20 μl of sample buffer. Eluted proteins were run on a 10% SDS/polyacrylamide gel and detected after Western blotting with the ECL® (enhanced chemiluminescence) system (GE Healthcare). Quantification of bands was carried out by densitometry. To this end, lanes of underexposed films (to stay as linear as possible) were profiled, and the area under the scan curve was used as an estimation of the co-precipitated protein amount compared with standards run in parallel. Results from a total of five independent experiments were averaged.
Monopolymerase system
All reactions contained in an end volume of 40 μl of 30 mM Hepes/KOH (pH 7.8), 7 mM magnesium acetate, 1 mM EGTA, 0.5 mM DTT (dithiothreitol), 40 mM phosphocreatine, 40 μg/μl creatine kinase, 4 mM ATP, 0.2 mM each of CTP, GTP and UTP, 0.2 mM each of dATP and dGTP, 0.05 mM each of dCTP and dTTP, 10 μCi each of [α-32P]dCTP and [α-32P]dTTP, 1.2 pmol of human Topo (0.1 μg/pmol), 3.5 pmol of RPA (0.116 μg/pmol), 6.25 pmol of SV40 Tag (0.096 μg/pmol), 1.1 pmol of human Pol (0.354 μg/pmol) and 0.1 pmol of pUC-HS DNA (2 μg/pmol) containing the complete SV40 origin sequences [23]. Since Pol will synthesize the leading and lagging strands in this type of reaction, the assay is referred to as the monopolymerase system [24]. For SV40 in vitro DNA replication, Topo, RPA and Pol were replaced by 190 μg of S100 extracts obtained from HEK-293 cells (human embryonic kidney cells) [25]. The sample was incubated for 90 min at 37 °C. A 5 μl volume of each reaction mixture was spotted on DE81 paper for quantification of incorporated labelled nucleotides [26]. The reactions were stopped by adding EDTA, SDS and proteinase K to final concentrations of 20 mM, 0.65% and 1.7 μg/μl respectively and a further 30 min of incubation. The sample was extracted once with phenol/chloroform, and DNA was passed over a G-50 spin column (Boehringer Mannheim) into TE buffer (10 mM Tris/HCl, pH 8, and 1 mM EDTA) to remove unincorporated nucleotides. DNA was ethanol-precipitated in the presence of 10 μg of carrier tRNA.
For the monopolymerase system, DNA was dissolved in 20 μl of alkaline loading buffer [50 mM NaOH, 1 mM EDTA, 5% (w/v) Ficoll 400 and 0.025% Bromcresol Green]. The samples were separated in 1.5% agarose gels in circulating alkaline running buffer (50 mM NaOH and 1mM EDTA) for 10 h at 150 mA in the cold. The gel was fixed in 10% (v/v) trichloroacetic acid, dried and exposed to X-ray films. For SV40 DNA replication, the DNA was dissolved in 20 μl of TE buffer, and 5 μl was double-restricted with EcoRI to linearize the plasmid DNA and DpnI to remove un-replicated DNA. Native loading buffer [20 mM Hepes/KOH, pH 8, 1 mM EDTA, 2% (w/v) sucrose and 0.01% Bromophenol Blue] was added, and products were separated in a 0.8% agarose gel in TBE (Tris/borate/EDTA; 89 mM Tris/borate, 89 mM boric acid and 10 mM EDTA). The gel was dried and exposed to X-ray films.
To analyse the effects of antibodies and peptides at different steps, ATP, other rNTPs and dNTPs were omitted in the initial sample to stall the monopolymerase reaction before the unwinding, primer synthesis and primer extension steps. Peptides or antibodies added to these reactions were pre-incubated with the proteins for 30 min prior to the release of the block by supplementing the missing nucleotides.
Enzymatic assays using natural ssDNA templates
Reactions were set up as for the monopolymerase system with two changes: first, Topo was omitted and, secondly, the SV40 origin containing DNA was replaced by 0.083 pmol of ssM13mp18 DNA (2.4 μg/pmol) or ssM13mp18 DNA primed with the universal primer oligonucleotide (5′-dGTAAAACGACGGCCAGT-3′; GE Healthcare) to serve as templates for primer synthesis and for primer extension respectively [27].
For primer synthesis, 0.2 mM each of GTP and UTP as well as 0.05 mM CTP along with 10 μCi of [α-32P]CTP were added. Extension reactions were supplemented instead with 0.2 mM each of dATP, dGTP and dTTP, 0.05 mM dCTP and 10 μCi of [α-32P]dCTP. Incubation proceeded for 90 min at 37 °C. Quantification and analysis of primer extension products were performed as described for the monopolymerase system. Products of the primer synthesis assays were ethanol-precipitated in the presence of 0.8 M LiCl, 10 mM MgCl2 and 10 μg of carrier tRNA. After dissolving the sample in 20 μl of denaturing loading buffer [35% (v/v) formamide, 8 mM EDTA, 0.1% Bromophenol Blue and 0.1% Xylene Cyanol FF] for 30 min at 65 °C, one-half of the samples were analysed in 20% denaturing urea/polyacrylamide gels in TBE. Autoradiography was performed with the wet gel.
RESULTS
Expression and characterization of peptides
We have expressed regions of Tag, Pol and RPA that are known to be involved in protein–protein interactions as soluble MBP-fusion peptides (see Supplementary Figure 1 at http://www.BiochemJ.org/bj/407/bj4070313add.htm). These included sequences spanning amino acids 164–249 for Tag (peptide T164-249) [9], amino acids 195–313 for the p180 subunit of Pol (peptide P195-313) [28] and amino acids 1–183 (peptide R1-173) and 174–250 (peptide R174-250) for the p70 subunit of RPA [8]. Peptides T164-249, P195-313 and R1-173 interacted specifically with the p32 and p70 subunits of RPA, Tag and the p180 subunit of Pol respectively. Peptide R174-250 bound to full-length Tag as well as the p48, p58 and p180 subunits of Pol (see Supplementary Figure 2 at http://www.BiochemJ.org/bj/407/bj4070313add.htm).
Since peptides associated specifically with certain replication factors, we next tested their ability to interfere with the interactions of the native proteins in co-precipitation assays.
Interference of peptides with protein–protein interactions
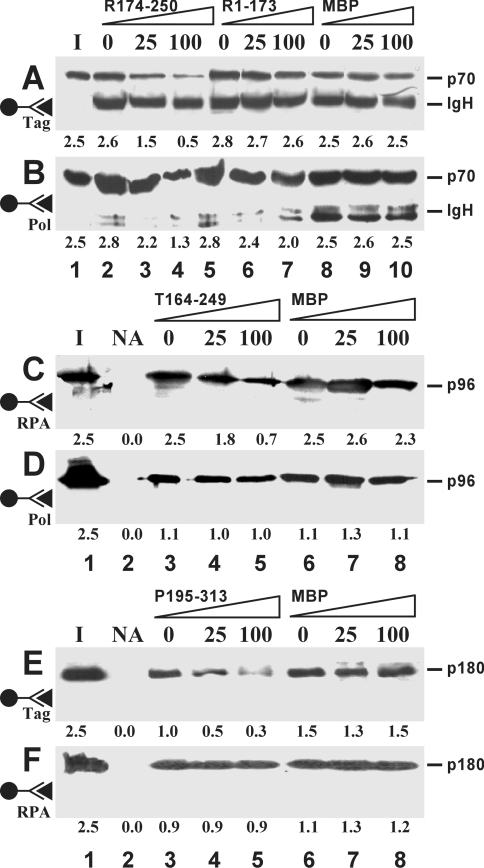
In the presence of peptides, specific associations between Tag, Pol and RPA were reduced in a dose-dependent manner (Figure 1). R174-250 diminished the amounts of RPA that were retained by either Tag (Figure 1A) or Pol (Figure 1B), whereas R1-173 slightly blocked the latter interaction. T164-249 specifically decreased the binding of Tag to RPA (Figure 1C), but left the interaction with Pol uninfluenced (Figure 1D). P195-313 inhibited the binding of Pol to Tag only (Figure 1E), but not that of Pol to RPA (Figure 1F). MBP on its own induced no reduction in these mutual interactions. The amounts of peptides needed to see a pronounced competition effect were in the range of a 100-fold molar excess and higher over the intact factors.
Figure 1. Inhibition of protein–protein interactions by peptides.
Pairwise co-precipitations were performed between Tag and RPA (A, C), Pol and RPA (B, F), and Pol and Tag (D, E), each at 25 pmol. The immobilized bait proteins are indicated by the antibody symbol on the left. Co-precipitated target proteins Tag (p96), Pol (p180 subunit) and RPA (p70 subunit) were revealed by Western blotting using the ECL® detection system. Peptides were present at a 25- and 100-fold molar excess over the target protein or were omitted (0). The numbers below each lane represent the average amount (in pmol) of co-precipitated protein as determined by densitometric scanning from five independent experiments. The position of the heavy chain of immunoglobulin (IgH) is indicated when it was detected due to cross-reactivity of the secondary antibodies. Lane I, one-tenth of the input material of soluble target proteins; lane NA, no antibody added.
We next investigated the effects the interference of protein–protein interactions exerted on DNA synthesis in the monopolymerase system.
Interference of peptides with the monopolymerase system
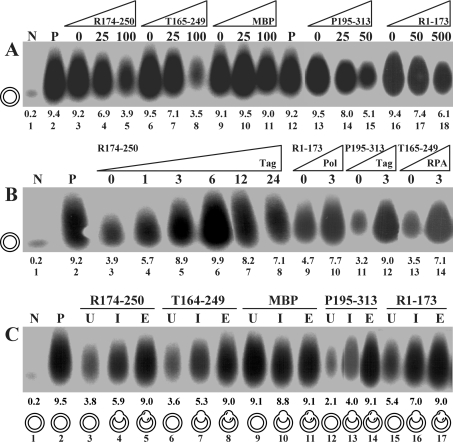
Peptides added at the beginning of monopolymerase reactions inhibited incorporation levels compared with the control reaction, depending on their concentration (Figure 2A). To see pronounced inhibition, a 50-, a 100- and a 500-fold excess over Tag were necessary for P195-313 (lanes 13–15), both T164-249 (lanes 6–8) and R174-250 (lanes 3–5), and R1-173 (lanes 16–18) respectively. MBP had no effect on the reaction at either of the concentrations used (lanes 9–11). In addition, we tested other MBP-fusion peptides negative for inhibition. These included fusions with the smaller RPA subunits (MBP-p14 and MBP-p32), a subfragment of the largest subunit spanning amino acids 330–440 (MBP-p70.330-440) and cyclin A (MBP-cyclin A) (results not shown). None of the peptides even at the highest concentrations used was able to diminish nucleotide incorporation to levels comparable with that of the negative control (lane 1). Back titration of up to 6 pmol of Tag into assays containing peptide R174-250 relieved inhibition, restoring incorporation to levels obtained in the absence of the peptide (Figure 2B, compare lanes 3–6 with lane 2). Increasing the amounts of Tag beyond 6 pmol led to a reduction of incorporation rates (lanes 7–8), suggesting quenching of other factors by this excess of Tag. Likewise, the inhibition of P195-313 and T164-249 could be reduced by addition of extra Tag (lanes 11–12) and RPA (lanes 13–14) respectively. Addition of the same amounts of these two factors to reactions without peptide showed only a minor stimulatory effect (results not shown). The addition of extra Pol increased incorporation levels in the presence of R1-173 (lanes 9–10), but stimulated the reaction to the same relative extent in the peptide's absence (results not shown).
Figure 2. Influence of peptides on the monopolymerase system.
(A) Titration of peptides. Increasing amounts of indicated peptides displayed as fold molar excess over 6.25 pmol of Tag were titrated into the reaction at the onset of monopolymerase reactions. (B) Back-titration of replication factors. Replication factors were added back at the indicated amounts at the onset of the reactions. Molar excess of peptides over Tag was 100-fold (for R1-173, 500-fold). (C) Kinetics of inhibition. Indicated peptides were added to the reaction mixture at a 100-fold (500-fold for R1-173) molar excess over Tag before unwinding (U), primer synthesis (I) and primer elongation (E). The different template configurations present at the time at which the peptides were administered were under-wound (U), primed (I) and primer-extended (E) plasmid DNAs, which are symbolized for each reaction. Lanes N and P represent negative and positive controls in the absence and presence of Tag. The numbers above lane numbers denote the amount of nucleotides incorporated (in pmol).
We next determined the time points during the monopolymerase reaction at which protein–protein interactions became important.
Influence of peptides on different steps of the monopolymerase system
Peptides were pre-incubated with stalled reactions prior to the unwinding, primer synthesis and primer extension steps by omitting ATP, other rNTPs and dNTPs from the reactions. All peptides were most potent if added prior to the unwinding step, when the template was a closed circular super-coiled substrate (Figure 2C, lanes labelled U). When the peptides were supplied after the unwinding step and before the onset of primer synthesis with a partially unwound template, intermediate inhibition levels were observed (lanes labelled I). When peptides were provided after the primer synthesis step and before the onset of the primer extension reaction, inhibition was almost completely lost and incorporation levels were comparable with that of the positive control (lanes labelled E). MBP showed no influence on the elongation rates regardless of when it was added to the reaction (lanes 9–11).
In an extension of these studies, we have also used other protocols to block the replication reactions at different time points (see Supplementary Methods and Supplementary Figure 3 at http://www.BiochemJ.org/bj/407/bj4070313add.htm). These results confirmed that the most pronounced inhibitory effect was seen as long as the peptides were administered prior to the primer synthesis step. Peptides added just prior to or after the primer extension reactions lost their inhibitory influence. The assembly of Tag on to the origin DNA and its bidirectional unwinding activities were affected by the peptides to a limited extent only.
The loss in interference at later replication steps could have been due to two reasons. First, the interactions between Tag, Pol and RPA were not essential for the primer extension step and following reactions, or, secondly, peptides could not bind to these factors any more once primer synthesis was started. To discriminate between these possibilities, we used monoclonal antibodies directed against the p70 subunit of RPA (70A, 70B and 70C) known to interfere with RPA's ability to interact with the other replicative factors (for a characterization of the antibodies, see Supplementary Figure 4 at http://www.BiochemJ.org/bj/407/bj4070313add.htm) [29]. In a first step, we tested the abilities of the antibodies to disrupt preformed complexes.
Interference of p70-specific monoclonal antibodies with protein–protein interactions
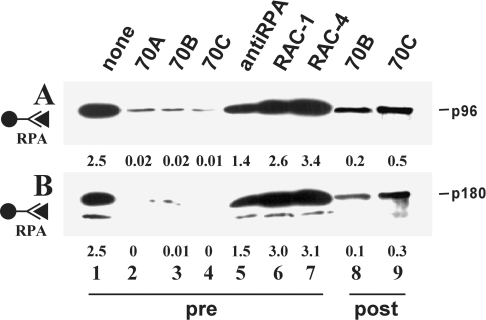
When antibodies 70A, 70B and 70C were pre-incubated with immobilized H6-RPA, its ability to interact with Tag and Pol was almost completely abolished (Figure 3, lanes 2–4). In contrast, RAC antibodies directed against the C-terminal part of p70 still allowed RPA to bind as efficiently as in the control reaction without antibodies (compare lanes 6–7 with lanes 1). A polyclonal serum proved somewhat inhibitory (lanes 5). When antibodies 70A, 70B and 70C were added to the reaction mixture after the replication factors were allowed to bind to each other, they efficiently disrupted the preformed interactions, and little of the soluble protein was retained by H6-RPA (shown for 70B and 70C in Figure 3, lanes 8–9).
Figure 3. Influence of p70-specific monoclonal antibodies on protein–protein interactions.
H6-RPA was linked to beads via anti-His antibodies. The resin was incubated with the indicated antibodies before (pre) or after (post) addition of the other replication factors. Anti-RPA refers to a polyclonal serum. Co-precipitated proteins were detected by Western blotting using the ECL® detection system. The positions of Tag (p96) and the p180 subunit of Pol are indicated. Lane 1 contained one-tenth of the input material of the soluble target protein. Numbers below each lane indicate the amounts of co-precipitated proteins (in pmol) as determined by densitometry.
We next tested the influence of antibodies on the different steps of the monopolymerase system.
Interference of p70-specific monoclonal antibodies with different steps of the monopolymerase reaction
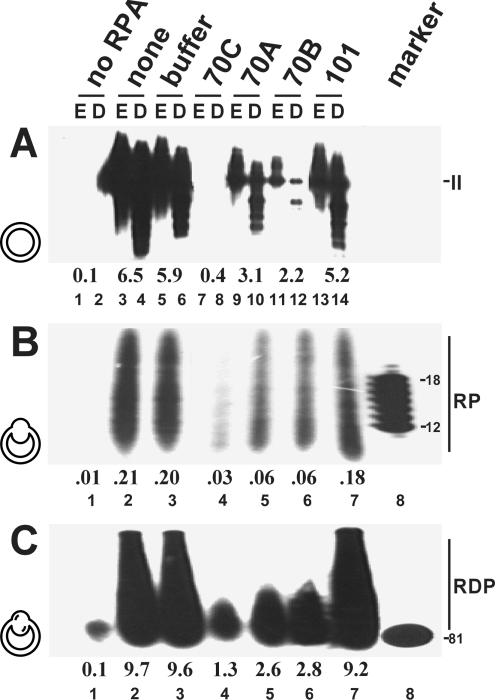
Antibodies 70A, 70B and 70C inhibited SV40 in vitro DNA replication (Figure 4A, lanes 7–12). Antibody 70C was most effective, abolishing the incorporation of nucleotides to levels comparable with that of the negative control without RPA (compare lane 4 with lane 1). Compared with positive controls (no antibody, lane 1; buffer, lane 2; and non-inhibitory Tag-specific antibody PAb 101, lane 7), antibodies 70A and 70B also significantly decreased incorporation levels.
Figure 4. Influence of p70-specific antibodies on replication-related activities of RPA.
Antibodies were tested in SV40 DNA replication (A), primer synthesis (B) and primer extension (C) assays. In (A), each DNA was restricted with EcoRI alone (E, odd-numbered lanes) or double- restricted by EcoRI–DpnI (D, even-numbered lanes). The numbers below the lanes indicate the amounts (in pmol) of incorporated dNMPs (A, C) or rNMPs (B). Lane 1 contained no protein. The positions of the products of each reaction, linear double-stranded DNA (form II), RNA primers (RP) and RNA–DNA primers (RDP), are indicated. Markers contained oligo(dT)8-12 or an 81-base-pair fragment. Antibodies were present at the onset of the reaction (A) or before the unwinding (B) or primer synthesis (C) steps. The template configurations present at the time at which antibodies were administered were super-coiled, under-wound and primed plasmid DNAs and are represented by symbols for each reaction.
When antibodies were supplied after the unwinding step and before starting primer synthesis with addition of rNTPs (Figure 4B) or after the priming step and before primer extension with the addition of dNTPs (Figure 4C) in monopolymerase assays, all three significantly reduced incorporation levels compared with the controls (compare lanes 4–6 with lanes 2,3 and 7). As was the case in the in vitro replication assay, 70C was the most powerful inhibitory antibody. Note that the antibody 70C required a 30 min pre-incubation period before the next reaction step was activated in order to exert strong inhibition, whereas both 70A and 70B inhibited without the requirement of a pronounced pre-incubation phase (results not shown).
To assess the significance of protein–protein interactions to a greater extent, antibodies were tested using un-primed and primed ssM13mp18 DNA as templates. These substrates do not require any unwinding step, making it possible to study the influence of antibodies directly on the stimulatory or inhibitory effects between the replication factors.
Influence of p70-specific monoclonal antibodies on RNA and DNA synthesis using natural single-stranded templates
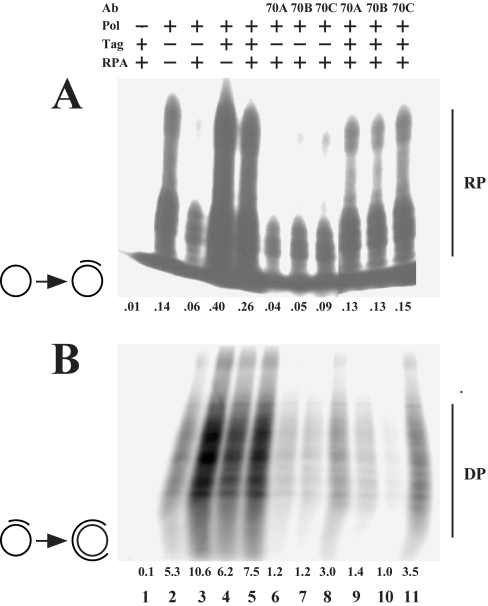
Using un-primed ssM13mp18 DNA as a template, no primer synthesis took place in the absence of Pol (Figure 5A, lane 1). In its presence, primers that were 2–25 nt in length were synthesized (lane 2). The presence of RPA inhibited primer synthesis (lane 3), whereas Tag led to an increase in the synthesis rate (lane 4). In addition, the inhibition induced by RPA was overcome in the presence of Tag (lane 5). In the absence of Tag, antibodies 70A and 70B left incorporation levels nearly uninfluenced; at most a slight decrease could be noticed (compare lanes 7 and 8 with lane 3). In contrast, 70C led to a slight stimulation of the primer synthesis reaction (compare lane 9 with lane 3). In the presence of Tag, all three antibodies reduced incorporation rates (compare lanes 9–11 with lane 5). Antibody 70C was hereby least efficient.
Figure 5. Influence of antibodies on primer synthesis and extension using natural ssDNA templates.
Primer synthesis (A) and primer extension (B) reactions were carried out using an un-primed or primed single-stranded circular DNA respectively as the templates, which are indicated by symbols for each reaction. The presence of any of the replication proteins Tag, Pol and RPA are indicated by ‘+’, their absence by ‘–’. Antibodies were added as specified (lanes 6–11). The numbers below each lane indicate the incorporated amounts (in pmol) of rNMPs (A) and dNMPs (B). RNA (RP) and DNA (DP) products are indicated.
Using a primed ssM13mp18 DNA template, Pol synthesized products 80–250 nt long (Figure 5B, lane 2). In its absence, no synthesis was observed (lane 1). In this setting, RPA and Tag stimulated polymerase activity, with RPA being more potent (lanes 3 and 4). Intermediate stimulation was seen in the presence of both proteins, indicating that Tag decreased the stimulatory effect of RPA to some extent (lane 5). In the absence or presence of Tag, all antibodies inhibited DNA synthesis, with 70C being significantly least efficient (compare lanes 6–8 and 9–11 with lane 3).
The influence of some of the interactions on enzymatic activities was also substantiated using the peptides (see Supplementary Figure 5 at http://www.BiochemJ.org/bj/407/bj4070313add.htm). Interference with the interaction between Tag and Pol by peptide P195-313 led to a decrease in the stimulatory effect of Tag on both the primer synthesis and primer extension reactions catalysed by Pol. Interfering with the Pol–RPA interaction led to a decrease in the efficiency of Pol to extend primers, but had no effect on the inhibition of primer synthesis exerted by RPA. Lastly, disturbing the interaction between Tag and RPA led to a decrease in Tag's ability to reverse the inhibitory and stimulatory effects of RPA on primer synthesis and primer extension respectively.
DISCUSSION
Replication of the SV40 mini-chromosome in cell-free extracts starts when the virally encoded Tag binds to the origin sequences as a double hexameric complex [3]. In subsequent steps, Tag marshals the association of three cellular factors, RPA [6,12], Pol [28,30] and Topo [11,31,32], to generate the initiation complex. After the initiation phase provides RNA–DNA primers for the leading and lagging strand DNA synthesis, full replication is accomplished by additional cellular factors [4]. In the present study, we investigated the significance of the physical interactions between Tag, Pol and RPA during different steps in the initiation reactions by using inhibitory peptides and antibodies.
The employed peptides and antibodies were able to interfere or disrupt specific interactions between Tag, RPA and Pol (Figures 1 and 3), but had otherwise no influence on replication activities of these factors (results not shown and Supplementary Figures 1 and 3). Only antibody 70C, which binds close to DBD-A of the p70 subunit of RPA, showed a destabilization of the RPA–DNA complex and, as a consequence, interfered with Tag-catalysed bidirectional unwinding, which needs an ssDNA-binding activity [8,29,33]. Antibodies 70A and 70B, which bind at the N-terminal part of RPA, were very potent in blocking the formation of protein–protein contacts and disrupted preformed complexes, probably due to steric hindrance. This possibility is consistent with the observation that both antibodies inhibit Pol stimulation by RPA [29]. Surprisingly, antibody 70C was reported not to interfere with Pol stimulation [29], albeit it interfered strongly with Pol binding in our hands. However, this antibody needed a prolonged pre-incubation step with RPA in order to exert its inhibitory effects in enzymatic assays, and therefore its negative influence might easily have escaped observation.
In contrast with antibodies, peptides were markedly selective in blocking association reactions (Figure 1). P195-313 and T164-249 interfered solely with Tag's and RPA's ability to interact with those partner molecules for which they mimicked the binding interface, which are Pol and Tag [9,28]. R1-173, on the other hand, interfered exclusively with the docking of Pol on RPA, consistent with the idea that the corresponding sequences on the p70 subunit are important for Pol stimulation [7,8]. R174-250, representing a sequence within DBD-A, blocked access of both Tag and Pol, emphasizing the role of this domain in the control of Pol's processivity, which is positively and negatively regulated by RPA and Tag respectively [8]. The presence of the peptides at the onset of the reaction inhibited the incorporation levels in the monopolymerase system in a concentration-dependent manner (Figure 2). The huge peptide excess over replication factors needed to exert significant inhibition indicate that the peptides in all probability do not contain the full set of amino acids involved in forming contact sites between the full-length proteins. In addition, some peptides such as R1-174 might interfere with the initial docking of the interacting molecules only, but might not block their major binding sites (see Supplementary Figure 2). The inhibitory effect of the peptides could be overcome by adding back the respective replication factor, corroborating the interpretation that interferences were due to disturbances in protein–protein interactions.
Kinetics of inhibition showed that the peptides were most effective if present before the unwinding step, which itself was only slightly affected. Inhibition capacity was reduced if peptides were added after unwinding and before primer synthesis and ceased completely when they were supplemented after the primer synthesis and before the primer extension reaction. This suggests that the peptides can form unproductive complexes with replication factors only during the early steps in the initiation reaction. Our results indicate that interactions were transient during the assembly step of Tag on to the origin DNA (Supplementary Figure 5). They became metastable during the unwinding reaction, when the pre-initiation complex is assembled. Once Pol was engaged in primer synthesis, a stable initiation complex or primosome was formed that acted in a processive manner. The fact that complex disruption by antibodies could still interfere with primer synthesis and extension at these late stages shows the importance of the physical interactions between Tag, RPA and Pol throughout the initiation process.
The significance of mutual interactions on enzymatic activities was additionally demonstrated in inhibition studies using natural ssDNA templates (Figure 5 and Supplementary Figure 5). Primer synthesis on circular ssDNA templates by Pol was greatly reduced in the presence of RPA. Interference with the RPA–Pol interaction did not relieve the block, suggesting that the inhibition is indirectly exerted by RPA by blocking Pol's access to the template [12,13,34]. Such an occlusion effect was corroborated by a slight stimulatory effect of antibody 70C, which destabilizes the DNA–RPA complex. Inhibition was overcome in the presence of Tag, but the latter's stimulatory effect was abrogated to a great extent or diminished by all antibodies and by those peptides that inhibited Tag's ability to bind to RPA. This suggests that Tag, in co-operation with Pol, can alter the DNA binding properties of RPA through direct interactions [35]. With respect to the antibodies, 70C was less potent in its inhibitory power than 70A and 70B, probably because the destabilizing influence on DNA binding reversed the negative effect exerted by the disruption of protein–protein interactions to a certain level. A primed template, in contrast, was a better substrate for Pol when coated with RPA, and a strong stimulation in the polymerization activity could be observed compared with a naked primed circular ssDNA substrate [7,8,19,36]. In this setting, RPA seemed to block unproductive binding sites on the template and to actively stabilize Pol on the primer–template junction, since all antibodies and peptides inhibiting the Pol–RPA interaction abolished the stimulatory effect to a great or some extent respectively. In addition, the clearance of RPA that is required during polymerization might be due to Pol–RPA interactions. Antibody 70C with its destabilizing effect on RPA–DNA complexes would facilitate the release of RPA, accounting further for its lower inhibition capacity compared with the other two inhibitory antibodies 70A and 70B. Tag was stimulatory to both the priming and polymerization activity of Pol using the naked primed template, underlining the importance of its interaction with the p180 subunit of Pol. Indeed, peptides inhibitory for the Pol–Tag interaction reduced this positive effect in both settings [15,28]. On the other hand, the presence of Tag reduced incorporation levels on the RPA-covered substrate. This might have been due to a stabilizing effect on the DNA binding of RPA, since antibody 70C could reverse this inhibition to a great extent and interference with the Tag–RPA interaction by peptide T164-249 also resulted in a stimulation of the extension reaction.
Experiments to explain the precise assembly of the initiation complex that culminates in the synthesis of RNA primers and their extension with DNA are ongoing [10,14]. Inhibition studies such as the one in the present paper should be helpful to understand the dynamics of the replication process in terms of the timed formation of protein–protein as well as protein–DNA interactions in future work.
Online data
Acknowledgments
This work was supported by the NIH (National Institutes of Health); (grants GM52948 to E. F. and C00004284-1 to H.-P. N.), Vanderbilt University (grant to E. F.), Deutsche Forschungsgemeinschaft (grant SFB604 to F. G.), Health Research Board Ireland (grant RP/2004/154 to H.-P. N.) and Science Foundation Ireland (grant 05/RFP/BIC0022 to H.-P. N.). The Leibniz Institute for Age Research is supported financially by the federal government and the Land Thueringen.
References
- 1.Kelly T. J. SV40 DNA replication. J. Biol. Chem. 1988;263:17889–17892. [PubMed] [Google Scholar]
- 2.Stenlund A. Initiation of DNA replication: lessons from viral initiator proteins. Nat. Rev. Mol. Cell Biol. 2003;4:777–785. doi: 10.1038/nrm1226. [DOI] [PubMed] [Google Scholar]
- 3.Simmons D. T. SV40 large T antigen functions in DNA replication and transformation. Adv. Virus Res. 2000;55:75–134. doi: 10.1016/s0065-3527(00)55002-7. [DOI] [PubMed] [Google Scholar]
- 4.Waga S., Bauer G., Stillman B. Reconstitution of complete SV40 DNA replication with purified replication factors. J. Biol. Chem. 1994;269:10923–10934. [PubMed] [Google Scholar]
- 5.Stauffer M. E., Chazin W. J. Structural mechanisms of DNA replication, repair, and recombination. J. Biol. Chem. 2004;279:30915–30918. doi: 10.1074/jbc.R400015200. [DOI] [PubMed] [Google Scholar]
- 6.Dornreiter I., Erdile L. F., Gilbert I. U., von Winkler D., Kelly T. J., Fanning E. Interaction of DNA polymerase α-primase with cellular replication protein A and SV40 T antigen. EMBO J. 1992;11:769–776. doi: 10.1002/j.1460-2075.1992.tb05110.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim D. K., Stigger E., Lee S. H. Role of the 70-kDa subunit of human replication protein A (I). Single-stranded DNA binding activity, but not polymerase stimulatory activity, is required for DNA replication. J. Biol. Chem. 1996;271:15124–15129. doi: 10.1074/jbc.271.25.15124. [DOI] [PubMed] [Google Scholar]
- 8.Braun K. A., Lao Y., He Z., Ingles C. J., Wold M. S. Role of protein–protein interactions in the function of replication protein A (RPA): RPA modulates the activity of DNA polymerase α by multiple mechanisms. Biochemistry. 1997;36:8443–8454. doi: 10.1021/bi970473r. [DOI] [PubMed] [Google Scholar]
- 9.Weisshart K., Taneja P., Fanning E. The replication protein A binding site in simian virus 40 (SV40) T antigen and its role in the initial steps of SV40 DNA replication. J. Virol. 1998;72:9771–9781. doi: 10.1128/jvi.72.12.9771-9781.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arunkumar A. I., Klimovich V., Jiang X., Ott R. D., Mizoue L., Fanning E., Chazin W. J. Insights into hRPA32 C-terminal domain-mediated assembly of the simian virus 40 replisome. Nat. Struct. Mol. Biol. 2005;12:332–339. doi: 10.1038/nsmbXX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roy R., Trowbridge P., Yang Z., Champoux J. J., Simmons D. T. The cap region of topoisomerase I binds to sites near both ends of simian virus 40 T antigen. J. Virol. 2003;77:9809–9816. doi: 10.1128/JVI.77.18.9809-9816.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Melendy T., Stillman B. An interaction between replication protein A and SV40 T antigen appears essential for primosome assembly during SV40 DNA replication. J. Biol. Chem. 1993;268:3389–3395. [PubMed] [Google Scholar]
- 13.Collins K. L., Kelly T. J. The effects of T antigen and replication protein A on the initiation of DNA synthesis by DNA polymerase α-primase. Mol. Cell. Biol. 1991;11:2108–2115. doi: 10.1128/mcb.11.4.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Simmons D. T., Gai D., Parsons R., Debes A., Roy R. Assembly of the replication initiation complex on SV40 origin DNA. Nucleic Acids Res. 2004;32:1103–1112. doi: 10.1093/nar/gkh236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dornreiter I., Hoss A., Arthur A. K., Fanning E. SV40 T antigen binds directly to the large subunit of purified DNA polymerase α. EMBO J. 1990;9:3329–3336. doi: 10.1002/j.1460-2075.1990.tb07533.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stadlbauer F., Brueckner A., Rehfuess C., Eckerskorn C., Lottspeich F., Förster V., Tseng B. Y., Nasheuer H. P. DNA replication in vitro by recombinant DNA-polymerase-α-primase. Eur. J. Biochem. 1994;222:781–793. doi: 10.1111/j.1432-1033.1994.tb18925.x. [DOI] [PubMed] [Google Scholar]
- 17.Soe K., Dianov G., Nasheuer H. P., Bohr V. A., Grosse F., Stevnsner T. A human topoisomerase I cleavage complex is recognized by an additional human topoisomerase I molecule in vitro. Nucleic Acids Res. 2001;29:3195–3203. doi: 10.1093/nar/29.15.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Henricksen L. A., Umbricht C. B., Wold M. S. Recombinant replication protein A: expression, complex formation, and functional characterization. J. Biol. Chem. 1994;269:11121–11132. [PubMed] [Google Scholar]
- 19.Kolpashchikov D. M., Weisshart K., Nasheuer H. P., Khodyreva S. N., Fanning E., Favre A., Lavrik O. I. Interaction of the p70 subunit of RPA with a DNA template directs p32 to the 3′-end of nascent DNA. FEBS Lett. 1999;450:131–134. doi: 10.1016/s0014-5793(99)00484-6. [DOI] [PubMed] [Google Scholar]
- 20.Weisshart K., Forster H., Kremmer E., Schlott B., Grosse F., Nasheuer H. P. Protein–protein interactions of the primase subunits p58 and p48 with simian virus 40 T antigen are required for efficient primer synthesis in a cell-free system. J. Biol. Chem. 2000;275:17328–17337. doi: 10.1074/jbc.M000717200. [DOI] [PubMed] [Google Scholar]
- 21.Gurney E. G., Harrison R. O., Fenno J. Monoclonal antibodies against simian virus 40 T antigens: evidence for distinct sublcasses of large T antigen and for similarities among nonviral T antigens. J. Virol. 1980;34:752–763. doi: 10.1128/jvi.34.3.752-763.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tanaka S., Hu S.-Z., Wang T. S.-F., Korn D. Preparation and preliminary characterization of monoclonal antibodies against human DNA polymerase α. J. Biol. Chem. 1982;257:8386–8390. [PubMed] [Google Scholar]
- 23.Weiner B. M., Bradley M. K. Specific mutation of a regulatory site within the ATP-binding region of simian virus 40 large T antigen. J. Virol. 1991;65:4973–4984. doi: 10.1128/jvi.65.9.4973-4984.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murakami Y., Hurwitz J. DNA polymerase α stimulates the ATP-dependent binding of simian virus tumor T antigen to the SV40 origin of replication. J. Biol. Chem. 1993;268:11018–11027. [PubMed] [Google Scholar]
- 25.Guo Z.-S., Gutierrez C., Heine U., Sogo J. M., DePamphilis M. L. Origin auxiliary sequences can facilitate initiation of simian virus 40 DNA replication in vitro as they do in vivo. Mol. Cell. Biol. 1989;9:3593–3602. doi: 10.1128/mcb.9.9.3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maniatis T., Fritsch E. F., Sambrook J. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 1982. Molecular Cloning: A Laboratory Manual. [Google Scholar]
- 27.Schneider C., Weisshart K., Guarino L. A., Dornreiter I., Fanning E. Species specific functional interactions of DNA polymerase α-primase with SV40 T antigen require SV40 origin DNA. Mol. Cell. Biol. 1994;14:3176–3185. doi: 10.1128/mcb.14.5.3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dornreiter I., Copeland W. C., Wang T. S. Initiation of simian virus 40 DNA replication requires the interaction of a specific domain of human DNA polymerase α with large T antigen. Mol. Cell. Biol. 1993;13:809–820. doi: 10.1128/mcb.13.2.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kenny M. K., Schlegel U., Furneaux H., Hurwitz J. The role of human single-stranded DNA binding protein and its individual subunits in simian virus 40 DNA replication. J. Biol. Chem. 1990;265:7693–7700. [PubMed] [Google Scholar]
- 30.Murakami Y., Hurwitz J. Functional interactions between SV40 T antigen and other replication proteins at the replication fork. J. Biol. Chem. 1993;268:11008–11017. [PubMed] [Google Scholar]
- 31.Trowbridge P. W., Roy R., Simmons D. T. Human topoisomerase I promotes initiation of simian virus 40 DNA replication in vitro. Mol. Cell. Biol. 1999;19:1686–1694. doi: 10.1128/mcb.19.3.1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seinsoth S., Uhlmann-Schiffler H., Stahl H. Bidirectional DNA unwinding by a ternary complex of T antigen, nucleolin and topoisomerase I. EMBO Rep. 2003;4:263–268. doi: 10.1038/sj.embor.embor770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brill S. J., Stillman B. Yeast replication factor-A functions in the unwinding of the SV40 origin of DNA replication. Nature. 1989;342:92–95. doi: 10.1038/342092a0. [DOI] [PubMed] [Google Scholar]
- 34.Murakami Y., Eki T., Hurwitz J. Studies on the initiation of simian virus 40 replication in vitro: RNA primer synthesis and its elongation. Proc. Natl. Acad. Sci. U.S.A. 1992;89:952–956. doi: 10.1073/pnas.89.3.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiang X., Klimovich V., Arunkumar A. I., Hysinger E. B., Wang Y., Ott R. D., Guler G. D., Weiner B., Chazin W. J., Fanning E. Structural mechanism of RPA loading on DNA during activation of a simple pre-replication complex. EMBO J. 2006;25:5516–5526. doi: 10.1038/sj.emboj.7601432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kolpashchikov D. M., Khodyreva S. N., Khlimankov D. Y., Wold M. S., Favre A., Lavrik O. I. Polarity of human replication protein A binding to DNA. Nucleic Acids Res. 2001;29:373–379. doi: 10.1093/nar/29.2.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.