Abstract
ZFHX1A is expressed in proliferating cells in the developing embryo, and in the present study we provide evidence that its expression is confined to proliferating cells through dependence on the Rb (retinoblastoma protein) family/E2F cell cycle pathway. Mutation of the Rb or E2F1 genes lead to induction of ZFHX1A mRNA, implying that the Rb–E2F1 repressor complex is important for repression of ZFHX1A. This repression is associated with recruitment of an E2F–Rb–histone deacetylase repressor complex to the promoter. A dominant-negative form of E2F1 inhibited ZFHX1A expression in p16INK4a(−) cells where Rb is constitutively hyperphosphorylated and inactive, suggesting that E2F can contribute to ZFHX1A transactivation in the absence of functional Rb. ZFHX1A is an E-box-binding transcription factor whose binding sites overlap with those bound by Snail1 and 2, and ZFHX1B/SIP1 (leading to at least partially overlapping function; for example, each of the proteins can repress E-cadherin expression). We found that expression of Snail1 and ZFHX1B/SIP1 is also regulated by E2Fs, but in contrast with ZFHX1A this regulation is Rb-family-independent. Snail2 expression was unaffected by either E2F or the Rb family. We propose that the differential effects of the Rb family/E2F pathway on expression of these E-box-binding proteins are important in maintaining their distinct patterns (and thus distinct functions) during embryogenesis.
Keywords: proliferation, retinoblastoma protein, ZFHX1A, zinc finger transcription factor
Abbreviations: cdk, cyclin-dependent kinase; ChIP, chromatin immunoprecipitation; CMV, cytomegalovirus; CNS, central nervous system; CtBP, C-terminal-binding protein; DB, DNA-binding domain; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; HDAC, histone deacetylase; IPTG, isopropyl β-D-thiogalactoside; MEF, mouse embryonic fibroblast; mER, mutant oestrogen receptor; OHT, tamoxifen; Rb, retinoblastoma protein; RT, reverse transcriptase; TGF-β, transforming growth factor-β; TKO, triple knockout
INTRODUCTION
ZFHX1A (δEF1, ZEB1, TCF8, Zfhep) and ZFHX1B/SIP1 constitute a family of zinc finger homeodomain transcription factors whose function is conserved from Drosophila to humans. In Drosophila only a single family member is present (ZHF-1) [1], which appears to have diverged into two members in vertebrates [2,3]. Using common zinc finger domains, these factors bind to the same set of E-box-like sequences at target genes [3], and these sites overlap with those bound by the Snail family [4,5]. ZFHX1A/B and Snail proteins can each repress transcription, at least in part, through recruitment of the CtBP (C-terminal-binding protein) co-repressor, which is a component of a larger repressor complex containing HDAC (histone deacetylase) and polycomb proteins [4–6]. Although ZFHX1A/B and Snail proteins appear to be expressed in different subsets of cells and at different developmental times, each of the proteins has been shown to repress E-cadherin (an epithelial marker), to be overexpressed in different cancers and to cause epithelial-to-mesenchymal transition [4,7–10], implying that they have at least partially overlapping functions in vivo.
ZFHX1A can also serve as a transcriptional co-activator [11–13]. Co-activator function results, at least in part, from association with the histone acetyl transferase p300, which acetylates histones and disrupts inhibitory nucleosomes [10,14]. p300 also acetylates other transcription factors, and it has been demonstrated that this acetylation inhibits CtBP binding [15], suggesting a model whereby recruitment of p300 switches ZFHX1A from a CtBP-dependent repressor to a co-activator by blocking CtBP binding. This association with p300 only appears to occur when a second co-activator such as SMAD [TGF-β (transforming growth factor β) superfamily signalling mediators], which also associates with and is dependent upon p300, is present [14,16,17]. The proposed role for ZFHX1A in transcriptional activation is then to stabilize a Smad–p300 complex at the promoter of target genes. Consistent with this connection to SMADs, a recent study demonstrated an important role for ZFHX1A in TGF-β-mediated smooth muscle cell gene transcription in vivo [18].
ZFHX1A is present in muscle and skeletal progenitors as well as proliferating regions of the CNS (central nervous system) and migrating cranial neural crest ([10] and references therein). Further, ZFHX1A is also present in articular, meniscal and growth plate cartilage in the adult, where it can repress expression of CD-RAP (cartilage-derived retinoic-acid-sensitive protein) [19]. Loss of ZFHX1A leads to skeletal defects including shortened limbs, skeletal curvature and fusions, as well as craniofacial and eye defects characteristic of impaired cranial neural crest (defects resembling those seen when later stage embryos are exposed to retinoic acid) [20–22]. A subset of embryos have dramatic CNS defects including failure of neural tube closure at both caudal and cranial ends, and exencephaly. Heterozygous mutation of ZFHX1A leads to posterior polymorphous corneal dystrophy, in which there is a pathological epithelization of the corneal endothelium [23].
It has been shown that ZFHX1A is expressed in proliferating cells in developing mice and in cell culture [2]. Furthermore, knocking down ZFHX1A expression inhibited proliferation of cells in culture [24], implying that ZFHX1A may have a role in cell proliferation. In the present study, we provide evidence that expression of ZFHX1A in proliferating cells is linked to its direct regulation by Rb (retinoblastoma protein) and E2F1.
EXPERIMENTAL
Cells and cell culture
Rb family TKO (triple knockout) MEFs (mouse embryonic fibroblasts) and control wild-type fibroblasts were from Dr T. Jacks and Dr J. Sage (Cancer Center, M.I.T., Cambridge, MA, U.S.A.). Three independent TKO and wild-type isolates were used with similar results. E2F1-null cells were from Dr D. Johnson (Department of Carcinogenesis, University of Texas MD Anderson Cancer Center, Smithville, TX, U.S.A.) and Rb heterozygous and null cells were from Dr G. Leone (Human Cancer Genetics, The Ohio State University, Columbus, OH, U.S.A.). U2OS cells expressing IPTG (isopropyl β-D-thiogalactoside)-inducible p16INK4a were described previously [34], as were the U2OS cells expressing both IPTG-inducible p16INK4a and mER–DB-E2F [36]. U2OS cells were cultured with 1 mM IPTG in the medium for either 1 or 3 days to induce p16INK4a, or with 100 nM OHT (tamoxifen) for 1 day to induce mER–DB-E2F expression. For combined treatments, cells were treated with IPTG for 1 day, and then OHT was added along with IPTG for an additional day.
RNA extraction and real-time PCR
Total cellular RNA was extracted using TRIzol® solution (Invitrogen). Using Primer3 (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3.cgi), primer sets were designed to generate 100–200 bp PCR products that bridged two separate exons. Primer location and sequence, Tm (melting temperature) and PCR product sizes are listed in Supplementary Table 1 (see http://www.BiochemJ.org/bj/408/bj4080079add.htm). First-strand cDNA synthesis was carried out in 20 μl reaction volumes containing 5 μg of total RNA, 500 ng of random hexamers, 10 mM dithiothreitol, 500 μM dNTP mix, 40 units of RNaseOUT™ ribonuclease inhibitor and 200 units of M-MLV RT (reverse transcriptase) at 37 °C for 1 h according to the manufacturer's protocol (Invitrogen). Real-time quantitative PCR was performed in 25 μl reaction volumes containing 0.25 μl aliquots of cDNA, gene-specific primer pairs and SYBR Green I fluorescent dye (Molecular Probes) in an Mx3000P Real-Time PCR System (Stratagene), according to the manufacturer's protocol. The PCR cycle parameters were set at 95 °C for 20 s, 60 °C for 30 s and 72 °C for 30 s, for a total of no more than 45 cycles. The fluorescent intensity of SYBR Green was monitored at the end of each extension step; relative amounts of the target cDNA were estimated by the Ct (threshold cycle) number, and compared with two control genes, β-actin and GAPDH (glyceraldehyde-3-phosphate dehydrogenase). Three independent samples were analysed for each condition and/or cell type, and each sample was compared in at least three independent RT-PCR amplifications.
Transfection assays
The ZFHX1A promoter was amplified by PCR of human genomic DNA with the Expand High Fidelity PCR System (Roche) with a specific upstream primer (5′-GTGGGGTGGGGTCAATTCCATAGTC) and a primer within exon 1 (5′-TCCGCCATGATCCTCTCGCTTGT). A fragment of the promoter from −913 to +44 was cloned into pGL3-luciferase (Z1p.1000Luc) and 5′ truncations were created, Z1p.359Luc, Z1p.212Luc and Z1p.133Luc, by standard cloning methods. All constructs were sequenced to make sure that errors did not occur during the PCR amplification process. U2OS cells stably expressing OHT-inducible mER–DB-E2F were used for transient transfections to directly test ZFHX1A promoter activity. The cells were plated at 1.5×105 cells per well in 12-well plates, and transfected with Lipofectamine™ 2000 (Invitrogen) in serum-free medium. Transfections were performed in triplicate, and included 400 ng of the human ZFHX1A promoter-luciferase construct, and 100 ng of CMV (cytomegalovirus)-β-galactosidase DNA (Clontech) as a control for transfection efficiency. Where indicated, 100 mM OHT was added the next morning. At 2 days after transfection, cellular lysates were used for luciferase and β-galactosidase assays. Experiments were pooled and differences between groups were tested by ANOVA followed by a Student's t test to identify significant differences.
ChIP (chromatin immunoprecipitation) assay
ChIP assays were based on the UpState protocol (http://www.upstate.com/misc/protocol) using formaldehyde to crosslink genomic DNA. The chromatin was sheared to an average length of 300–500 bp. Monoclonal antibodies against Rb (Santa Cruz sc-50), E2F1 (Santa Cruz sc-193), E2F4 (Santa Cruz sc-866) and polyclonal antiserum against ZFHX1A [46] were used for immunoprecipitation. Equal amounts of anti-IgG or pre-immune serum were used as controls. The sequence and locations of primers (for ZFHX1A and GAPDH promoters and the ZFHX1A downstream sequence) and the expected size of the PCR products are shown in the Supplementary Tables at http://www.BiochemJ.org/bj/408/bj4080079add.htm. ChIP PCR reactions were similar to those described above for real-time PCR, but with additional 1% BSA and 1% DMSO, and the PCR programmes usually had a higher annealing temperature (e.g. 60–68 °C) and a longer extension time (e.g. 1 min).
RESULTS
Dependence of ZFHX1A expression on p16INK4a and E2F
ZFHX1A (and its Drosophila homologue ZFH-1) is expressed in proliferating progenitors that give rise to the skeleton and muscle, as well as the CNS and the eye, and this expression is lost as cells differentiate and become post-mitotic [1–3,10,22,24–29]. Defects have been observed in ZFHX1A-null mice in the developing skeleton, cornea and in the CNS (in a subset of the embryos). Knockdown of ZFHX1A led to inhibition of cell proliferation, suggesting a role for the protein in cell proliferation [24].
We wondered how ZFHX1A might be confined to proliferating cells. A key regulator of cell proliferation is the Rb family/E2F pathway (reviewed in [30,31]), thus we hypothesized that dependence on this pathway might be responsible for linking ZFHX1A expression to cell proliferation. Rb family members interact with DNA and histone-modifying enzymes to form repressor complexes which are targeted to genes through interaction with the E2F family of DNA-binding proteins [30–32]. These repressor complexes can block transactivation by E2Fs, and they trigger assembly of a heterochromatin-like conformation that further inhibits transcription [33].
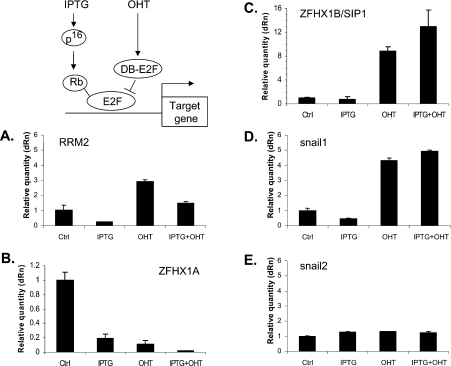
To determine whether ZFHX1A expression might be dependent upon Rb family members and E2Fs, we initially used a human Rb(+), p16INK4a(−) osteosarcoma cell line, U2OS, that expresses the cdk (cyclin-dependent kinase) inhibitor p16INK4a under the control of an IPTG-inducible promoter. We have described these cells previously [34]. p16INK4 blocks activity of cdk4 and 6, thereby specifically triggering accumulation of hypophosphorylated (active) Rb family members [35]. In the absence of IPTG and thus p16INK4a expression, Rb is constitutively hyperphosphorylated and inactive in these cells [34]. Treatment with IPTG led to inhibition of ZFHX1A expression, as it did with a known control cell cycle target gene, ribonucleotide reductase (Figures 1A and 1B). These results demonstrate that ZFHX1A expression is repressed when the Rb family is activated in response to p16INK4a in these cells.
Figure 1. Effects of p16INK4a and E2F binding on expression of ZFHX1A/B and Snail1/2 family members.
U2OS cells stably expressing IPTG-inducible p16INK4a and OHT-dependent mER–DB-E2F (where DB is a DNA-binding domain) were treated with IPTG, OHT or IPTG + OHT as described in the Experimental section. mRNA expression was followed by real-time PCR and compared with untreated cells (Ctrl). Results were normalized to β-actin mRNA. Similar results were seen with normalization to GAPDH mRNA. RRM2, ribonucleotide reductase.
To address a potential role for E2F, we used a dominant-negative-like form of E2F1 containing a DNA-binding domain (termed DB) but lacking transactivation and Rb-family-binding domains (Figure 1). We have used E2F-DB to displace E2F and Rb family–E2F complexes from genes previously [34], and the use of this construct has been reviewed recently [32]. To create this construct, E2F-1-DB was fused to a modified OHT-dependent oestrogen receptor (termed mER), and stably co-expressed into U2OS cells along with the IPTG-inducible p16INK4a vector. This cell system has been described in detail previously [36]. Upon treatment with OHT, the mER–DB-E2F fusion protein translocates from the cytoplasm to the nucleus, where it efficiently displaces E2Fs from target genes, based on ChIP assays [36]. As a control, treatment of the parent cells in this system (cells not expressing mER–DB-E2F) with OHT did not lead to significant changes in gene expression [36].
Treatment of cells with OHT activated the ribonucleotide reductase gene, and it also reversed the repression of ribonucleotide reductase seen when p16INK4a was induced by IPTG (Figure 1A). By contrast, we found that OHT inhibited ZFHX1A expression and repression by p16INK4a was not reversed by OHT treatment (Figure 1B).
Snail1 and ZFHX1B/SIP1 are repressed by E2F, but are unaffected by p16INK4a expression
Because ZFHX1B/SIP1, Snail1 and Snail2 (slug) bind similar E-box-like sequences and appear to have at least partially overlapping functions with ZFHX1A, we investigated whether these genes were also dependent upon p16INK4a and E2F. We found no effect of p16INK4a expression or activation of mER–DB-E2F on the expression of Snail2 (Figure 1E). However, both ZFHX1B and Snail1 were induced by mER–DB-E2F (Figures 1C and 1D), suggesting that, as with ribonucleotide reductase, E2F is participating in repression of these genes. However, expression of these genes was unaffected by p16INK4a expression. Thus Snail1 and ZFHX1B/SIP1 are regulated by E2F but they appear to be unaffected by Rb family member activation.
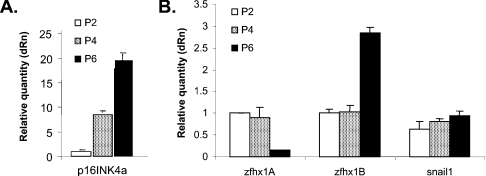
Induction of p16INK4a and repression of ZFHX1A occurs sequentially as MEFs are passaged in culture
As MEFs are passaged in culture, they express p16INK4a and ARF (alternate reading frame protein) from the INK4a locus [35,37]. Expression of these proteins leads to accumulation of hypophosphorylated Rb family members, and with time the cells undergo a permanent arrest (senescence) [33,37]. We compared the time course of p16INK4a mRNA expression with that of ZFHX1A during passage (denoted P) of MEFs in culture. p16INK4a mRNA was low at P2; however, it was significantly induced by P4 and further induced at P6 (Figure 2A). By contrast, ZFHX1A mRNA expression remained unchanged until P6, when it was repressed. Consistent with their lack of sensitivity to p16INK4a expression above, Snail1 and 2 expression did not change with passage number (and ZFHX1B expression was moderately induced at P6) (Figure 2B). These results are consistent with the notion that ZFHX1A is repressed following p16INK4a expression (as above when p16INK4a was overexpressed in U2OS cells).
Figure 2. Expression of p16INK4a precedes repression of ZFHX1A during passage of MEFs in culture; Snail1/2 and ZFHX1B/SIP1 expression is not repressed.
Real-time PCR analysis was performed to compare expression of the indicated mRNAs in MEFs at P2, 4 and 6 in culture. Expression was normalized to β-actin. Similar results were seen with normalization to GAPDH.
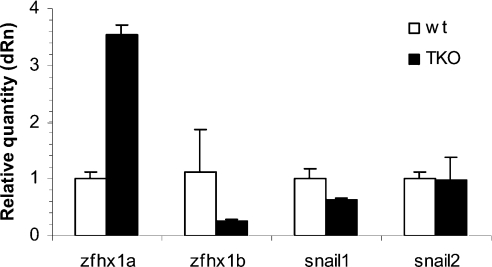
Mutation of the Rb family leads to induction of ZFHX1A
To examine the role of the Rb family more specifically, we used a second model system; MEFs derived from mice where all three Rb family member genes had been mutated (TKOs) [38]. Consistent with an inhibitory role for Rb family members in ZFHX1A expression (e.g. repression when p16INK4a was induced above), the mRNA was induced in the TKO MEFs compared with wild-type cells (Figure 3). By contrast, expression of ZFHX1B/SIP1 and Snail1 mRNA was not induced, consistent with the findings above that p16INK4a expression did not lead to repression of these genes (Figures 1 and 2).
Figure 3. Mutation of Rb family members leads to induction of ZFHX1A, but not ZFHX1B or Snail family members.
Gene expression was analysed in wild-type (wt) and TKO MEFs by real-time PCR. TKO cells were at P7 and wt cells were at P2. Results were normalized to β-actin expression. Similar results were seen on normalization to GAPDH.
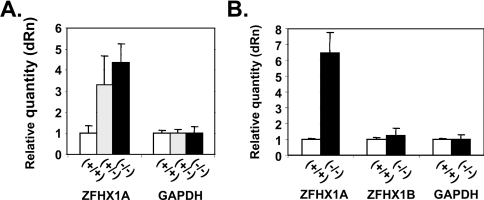
Rb and E2F1 participate in repression of ZFHX1A
We investigated whether Rb specifically was important for regulation of ZFHX1A expression. MEFs in which one or both Rb genes were mutated were compared with wild-type cells for expression of ZFHX1A mRNA using real-time PCR (Figure 4A). We found that mutation of a single copy of the Rb gene was sufficient for induction of ZFHX1A mRNA.
Figure 4. ZFHX1A expression is repressed by Rb and E2F1.
MEFs heterozygous or null for Rb, or null for E2F1 were compared with wild-type MEFs for expression of ZFHX1A mRNA using real-time PCR. (A) Rb mutant cells. (B) E2F1 mutant cells. Results were normalized to GAPDH mRNA. Similar results were seen with normalization to β-actin mRNA.
Next, we investigated whether E2F1 was important for regulation of ZFHX1A expression. MEFs in which both E2F1 genes were mutated were compared with wild-type control cells for ZFHX1A mRNA expression. As with the Rb mutation, we found induction of ZFHX1A mRNA in the E2F1-null cells (Figure 4B).
The ZFHX1A promoter is dependent upon E2F in transfection assays
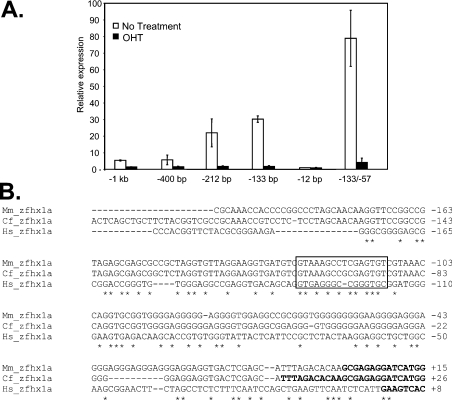
To investigate whether the ZFHX1A promoter was dependent upon E2F, we initially transfected a series of promoter deletion constructs into the U2OS cells stably expressing mER–DB-E2F, and measured transcription in the presence or absence of OHT (to activate this dominant-negative E2F). Deletion from −400 to −212 bp significantly activated the promoter, suggesting removal of a negative element (Figure 5A). However, promoter activity in the presence or absence of this negative element was inhibited by OHT, hence E2F binding appeared to be required for promoter activity. This OHT dependence was maintained upon deletion to −133 bp; however, OHT dependence was lost upon further deletion to −12 bp, which leaves only the TATA box. Next, we deleted the GC-rich region between −12 and −57 bp leaving the sequence between −57 and −133 bp upstream of the TATA box. This construct remained OHT-dependent (Figure 5A), demonstrating that the sequence between −57 and −133 bp is sufficient for responsiveness. The E2F-dependent sequence is located between −57 and −133 bp.
Figure 5. The ZFHX1A promoter is regulated by E2F binding in transfection assays.
(A) A series of ZFHX1A gene promoter constructs fused to the firefly luciferase gene were transfected into U2OS cells stably expressing mER–DB-E2F. Numbers in the construct name indicate base pairs of the 5-flanking sequence upstream of the ZFHX1A gene. The −133/−57 construct contains the sequence between −133 and −57 bp located upstream of the TATA box (−12 bp). Cells were treated with OHT to activate mER–DB-E2F. CMV-β-galactosidase was co-transfected in all wells and used to normalize the results. (B) ZFHX1A promoter sequence comparison in human, mouse and dog. The boxed region shows a conserved E2F-like consensus site within the responsive region.
Comparison of the mouse, dog and human ZFHX1A promoter sequences revealed a conserved consensus E2F-like-binding site within the responsive region (Figure 5B). Additional E2F-like consensus sequences were also evident further upstream in each of the species. However, it is of note that E2F sites are difficult to identify in vivo using a classic consensus sequence. Indeed, it was found that only 5% of such consensus sequences in the genome are bound to E2F1 and, perhaps more importantly, only 12% of sites bound by E2F1 show this consensus sequence [39].
The ZFHX1A gene binds E2F, Rb and HDAC in vivo
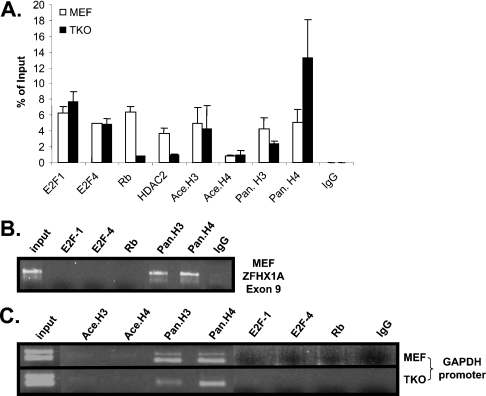
Because of the difficulty in identifying E2F-binding sites in vivo on the basis of sequence prediction and more importantly to determine whether the ZFHX1A promoter actually binds E2F and Rb in vivo, we examined the ZFHX1A promoter using ChIP assays. Two E2F family members, E2F1 and E2F4, were tested. We used PCR primers for the ChIP assays that spanned the promoter region between −133 and −57 bp. Both E2F1 and E2F4 were detected at the ZFHX1A promoter in wild-type cells (Figure 6A). Interestingly, both E2Fs were also detected at the promoter in TKO cells. E2F4 lacks a nuclear localization signal, and previously it has been thought that its appearance in the nucleus was dependent upon the interaction with an Rb family member [40]. However, recent studies demonstrate that E2F4 is present at a number of genes in the absence of Rb family members [41]. Rb was also detected at the promoter in wild-type cells, but not in TKOs (Figure 6A). As negative controls, no E2F or Rb binding was detected at the GAPDH gene or at a sequence downstream of the promoter within the ZFHX1A gene (Figures 6B and 6C). As a positive control, histones H3 and H4 were detected at these control sequences.
Figure 6. E2Fs bind the ZFHX1A promoter in vivo, and Rb and HDAC2 are recruited to the promoter in wild-type MEFs but not TKOs.
(A) Real-time PCR was used to quantify ChIP assays at the ZFHX1A promoter. Antibodies used for immunoprecipitation are shown at the bottom. (B and C) Gels following real-time PCR amplification are shown for controls: the GAPDH promoter and an internal sequence within the ZFHX1A gene. ‘Pan’ indicates antibodies recognizing all forms of histone H3 and H4, and ‘ace’ indicates antibodies specific for acetylated histones. IgG, control antibody.
It has been demonstrated previously that the Rb family can assemble repressor complexes including HDACs (reviewed in [31]), which remove inhibitory acetyl groups from histones, allowing them to assemble into transcriptionally repressive nucleosomes. We found that HDAC2 was recruited along with Rb and E2F to the ZFHX1A promoter in wild-type cells but not in TKOs (Figure 6A), consistent with the formation of an E2F–Rb–HDAC repressor complex at the ZFHX1A promoter in wild-type cells. Next, we investigated whether there was an increase in histone acetylation at the ZFHX1A promoter in the TKO cells, where Rb and HDAC2 are not recruited. We did not observe any change in overall acetylation of histone H3 or H4 at the promoter (Figure 6A). However, recruitment of HDAC may only affect histone acetylation in the immediate region of recruitment, such that global histone acetylation changes are not evident throughout the promoter. For example, at the cyclin E promoter, recruitment of Rb–HDAC has been shown to mediate repression by affecting histone acetylation at only a single adjacent nucleosome [42].
DISCUSSION
New studies are expanding Rb family/E2F target genes beyond the original set of cell cycle genes. Moreover, the roles for Rb family–E2F complexes appear to vary at these target genes. Generally, there are several categories of genes (probably a number of more complexities as well) [41]. One category consists of genes that are only repressed by Rb family–E2F when cells are growth-arrested by p16INK4a expression or upon serum starvation. A second and surprising category consists of genes which are constitutively repressed in a cell-cycle-independent fashion (no further repression of these genes is evident when p16INK4a is expressed or upon serum starvation). Another category consists of genes that are partially repressed by Rb family–E2F in proliferating cells, but are further repressed on cell arrest (e.g. via p16INK4a expression or serum starvation). It appears that ZFHX1A falls into this third category. Although its expression is repressed upon expression of p16INK4a, it is also induced in proliferating cells by Rb family mutation.
We suggest that an Rb–E2F1–HDAC repressor complex is important for repression of ZFHX1A. Additionally, in the absence of functional Rb [e.g. in the p16INK4a(−) U2OS cells] it appears that E2Fs can contribute to transactivation of ZFHX1A. However, as discussed above, it appears that assembly of Rb-family-dependent repressor complexes is not confined solely to arrested cells, and indeed we see evidence of ZFHX1A induction when Rb is mutated in proliferating cells. It is of note that acutely inactivating E2F activity with DB-E2F leads to down regulation of ZFHX1A in human U2OS cells, whereas ZFHX1A expression was induced in E2F1 mutant mouse cells. Therefore it is also possible that there may have been compensation from loss of E2F1 from another E2F family member in the E2F1 mutant cells, or these observations may reflect differences between mouse and human cells. p16INK4a expression is induced as mice age, and this is associated with senescence of stem cells in various tissues [43–45]. In a fashion analogous to p16INK4a, other cdks are important in regulating Rb family hyperphosphorylation to trigger the cell cycle arrest classically seen as progenitor cells undergo differentiation in vivo [35,37]. Thus other cdk inhibitors may be crucial in controlling ZFHX1A expression during embryonic development.
Interestingly, in contrast with ZFHX1A, Snail1 and ZFHX1B/SIP1 are repressed by E2F, and they are unaffected by the Rb family. Although E2Fs 1–5 can bind Rb family members, E2Fs 6–8 appear to be Rb-family-independent repressors (reviewed in [40]). Therefore it is possible that Snail1 and ZFHX1B might be repressed by such Rb-family-independent E2Fs. Alternatively, the effects on Snail1 and ZFHX1B may be indirect. Previously, a microarray-based assay was used to identify genes directly regulated by E2F (e.g. genes regulated when mER–DB-E2F was activated by OHT in the presence of CHX) [36]. Neither Snail1 nor ZFHX1B/SIP1 was identified as a direct E2F target in these studies, however, ZFHX1A was. Although these are negative results regarding Snail1 and ZFHX1B/SIP1, they may point toward an indirect mechanism for E2F in the regulation of the genes. Nevertheless, the differential dependence of the Snail1/2 and ZFHX1A/B genes on Rb family–E2F is interesting, and it may have implications for dictating the patterns of expression of these genes. Establishing distinct patterns for these genes may be particularly important because they bind a similar set of E-boxes, and they are also each dependent upon on the same co-repressor, CtBP. Taken together, these results imply that the four proteins are capable of targeting a similar subset of genes (e.g. E-cadherin) and perhaps repressing them through a similar mechanism (CtBP). Thus their unique functions in vivo may be dependent significantly on when and where they are expressed during development.
Online data
Acknowledgments
We thank Dr T. Jacks and Dr J. Sage for the gift of TKO and wild-type litter-mate control MEFs, Dr G. Leone for Rb heterozygous and null cells, and Dr D. Johnson for E2F1-null cells. The studies were supported in part by grants from the NIH (National Institutes of Health) to D. C. D. and D. S. D., and by NIH Center for Biomedical Research Excellence in Molecular Targets Grant (5P20 RR 018733).
References
- 1.Lai Z. C., Rushton E., Bate M., Rubin G. M. Loss of function of the Drosophila zfh-1 gene results in abnormal development of mesodermally derived tissues. Proc. Natl. Acad. Sci. U.S.A. 1993;90:4122–4126. doi: 10.1073/pnas.90.9.4122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Darling D. S., Stearman R. P., Qi Y., Qiu M. S., Feller J. P. Expression of Zfhep/δEf1 protein in palate, neural progenitors and differentiated neurons. Gene Expr. Patterns. 2003;3:709–717. doi: 10.1016/s1567-133x(03)00147-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Postigo A. A., Dean D. C. Differential expression and function of members of the zfh-1 family of zinc finger/homeodomain repressors. Proc. Natl Acad. Sci. U.S.A. 2000;97:6391–6396. doi: 10.1073/pnas.97.12.6391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hemavathy K., Ashraf S. L., Ip Y. T. Snail/slug family of repressors: slowly going into the fast lane of development and cancer. Gene. 2005;257:1–12. doi: 10.1016/s0378-1119(00)00371-1. [DOI] [PubMed] [Google Scholar]
- 5.Postigo A. A., Dean D. C. ZEB represses transcription through interaction with the corepressor CtBP. Proc. Natl. Acad. Sci. U.S.A. 1999;96:6683–6688. doi: 10.1073/pnas.96.12.6683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grooteclaes M. L., Frisch S. M. Evidence for a function of CtBP in epithelial gene regulation and anoikis. Oncogene. 2000;19:3823–3828. doi: 10.1038/sj.onc.1203721. [DOI] [PubMed] [Google Scholar]
- 7.Comijn J., Berx G., Vermassen P., Verschueren K., van Grunsven L., Bruyneel E., Mareel M., Huylebroeck D., van Roy F. The two-handed E box binding zinc finger protein SIP1 downregulates E-cadherin and induces invasion. Mol. Cell. 2001;7:1267–1278. doi: 10.1016/s1097-2765(01)00260-x. [DOI] [PubMed] [Google Scholar]
- 8.Eger A., Aigner K., Sonderegger S., Dampier B., Oehler S., Schreiber M., Berx G., Cano A., Foisner R. δEf1 is a transcriptional repressor of e-cadherin and regulates epithelial plasticity in breast cancer cells. Oncogene. 2005;24:2375–2385. doi: 10.1038/sj.onc.1208429. [DOI] [PubMed] [Google Scholar]
- 9.Spoelstra N. S., Manning N. G., Higashi Y., Darling D., Singh M., Shroyer K. R., Broaddus R. R., Horwitz K. B., Richer J. K. The transcription factor ZEB1 is aberrantly expressed in aggressive uterine cancers. Cancer Res. 2006;66:3893–3902. doi: 10.1158/0008-5472.CAN-05-2881. [DOI] [PubMed] [Google Scholar]
- 10.van Grunsven L. A., Tealman V., Michiels C., Opdecamp L., Huylebroeck D., Bellefroid E. J. δEF1 and SIP1 are differentially expressed and have overlapping activities during Xenopus embryogenesis. Dev. Dyn. 2006;235:1491–1500. doi: 10.1002/dvdy.20727. [DOI] [PubMed] [Google Scholar]
- 11.Chamberlain E. M. M., Sanders M. Identification of the novel player δEF1 in estrogen transcriptional cascades. Mol. Cell. Biol. 1999;19:3600–3606. doi: 10.1128/mcb.19.5.3600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dillner N. B., Sanders M. M. The zinc finger/homeodomain protein δEF1 mediates estrogen-specific induction of the ovalbumin gene. Mol. Cell. Endocrinol. 2002;192:85–91. doi: 10.1016/s0303-7207(02)00088-6. [DOI] [PubMed] [Google Scholar]
- 13.Lazarova D. L., Bordonaro M., Sartorelli A. C. Transcriptional regulation of the vitamin D3 receptor gene by ZEB. Cell Growth Differ. 2001;12:319–326. [PubMed] [Google Scholar]
- 14.Postigo A., Depp J. L., Taylor J. T., Kroll K. L. Regulation of Smad signaling through a differential recruitment of coactivators and corepressors by ZEB proteins. EMBO J. 2003;22:2453–2462. doi: 10.1093/emboj/cdg226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Q., Yao H., Vo N., Goodman R. H. Acetylation of adenovirus E1a regulates binding of the transcriptional corepressors CtBP. Proc. Natl. Acad. Sci. U.S.A. 2000;97:14323–14328. doi: 10.1073/pnas.011283598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Attisano L., Wrana J. L. Smads as transcriptional modulators. Curr. Opin. Cell Biol. 2000;12:235–243. doi: 10.1016/s0955-0674(99)00081-2. [DOI] [PubMed] [Google Scholar]
- 17.Postigo A. A. Opposing functions of ZEB proteins in the regulation of the TGFβ/BMP signaling pathway. EMBO J. 2003;22:2443–2452. doi: 10.1093/emboj/cdg225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nishimura G., Manabe I., Tsushima K., Maemura K., Makoto M., Higashi Y., Kondoh H., Nagai R. δEF1 regulates TGF-β signaling in vascular smooth muscle cell differentiation. Dev. Cell. 2006;11:93–104. doi: 10.1016/j.devcel.2006.05.011. [DOI] [PubMed] [Google Scholar]
- 19.Davies S. R., Sakano S., Zhu Y., Sandell L. J. Distribution of the transcription factors Sox9, AP-2, and δEF1 in adult murine articular and meniscal cartilage and growth plate. J. Histochem. Cytochem. 2002;50:1059–1065. doi: 10.1177/002215540205000808. [DOI] [PubMed] [Google Scholar]
- 20.Ross S. A., McCaffery P. J., Dragar U. C., DeLuca L. M. Retinoids in embryonal development. Physiol. Rev. 2000;80:1021–1054. doi: 10.1152/physrev.2000.80.3.1021. [DOI] [PubMed] [Google Scholar]
- 21.Higashi Y., Moribe H., Takagi T., Sekido R., Kawakami K., Kikutani H., Kondoh H. Impairment of T cell development in δEf1 mutant mice. J. Exp. Med. 1997;185:1467–1479. doi: 10.1084/jem.185.8.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takagi T., Moribe H., Kondoh H., Higashi Y. δEF1, a zinc finger and homeodomain transcription factor, is required for skeleton patterning in multiple lineages. Development. 1998;125:21–31. doi: 10.1242/dev.125.1.21. [DOI] [PubMed] [Google Scholar]
- 23.Krafchak C. M., Pawar H., Moroi S. E., Sugar A., Lichter P. R., Mackey D. A., Mian S., Narius T., Elner V., Schteingart M. T., et al. Mutations in TCF8 cause posterior polymorphous corneal dystrophy and ectopic expression of COL4A3 by corneal endothelial cells. Am. J. Hum. Genet. 2005;77:694–708. doi: 10.1086/497348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yen G., Crocia S., Dowling A., Zhang S., Zoeller R. T., Darling D. S. Developmental and functional evidence of a role for Zfhep in neural cell development. Brain Res. Mol. Brain Res. 2001;96:59–67. doi: 10.1016/s0169-328x(01)00267-4. [DOI] [PubMed] [Google Scholar]
- 25.Murray D., Precht P., Balakir R., Horton W. E. The transcription factor δEF1 is inversely expressed with Type II collagen mRNA and can repress Col2a1 promoter activity in transfected chondrocytes. J. Biol. Chem. 2000;275:3610–3618. doi: 10.1074/jbc.275.5.3610. [DOI] [PubMed] [Google Scholar]
- 26.Postigo A. P., Dean D. C. ZEB, a vertebrate homolog of Drosophila Zfh-1, is a negative regulator of muscle differentiation. EMBO J. 1997;16:3935–3943. doi: 10.1093/emboj/16.13.3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Postigo A. P., Ward E., Skeath J. B., Dean D. C. zfh-1, the Drosophila homologue of ZEB, is a transcriptional repressor that regulates somatic myogenesis. Mol. Cell. Biol. 1999;19:7255–7263. doi: 10.1128/mcb.19.10.7255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sooy K., Demay M. B. Transcriptional repression of the rat osteocalcin gene by δEF1. Endocrinology. 2002;143:3370–3375. doi: 10.1210/en.2001-211441. [DOI] [PubMed] [Google Scholar]
- 29.Terraz C., Toman D., Delauche M., Ronco P., Rossert J. A. δEF1 binds to a far-upstream sequence of the pro α1(I) collagen gene and represses its expression in osteoblasts. J. Biol. Chem. 2001;276:37011–37019. doi: 10.1074/jbc.M104185200. [DOI] [PubMed] [Google Scholar]
- 30.Frolov M. V., Dyson N. J. Molecular mechanisms of E2F-dependent activation and pRB-mediated repression. J. Cell Sci. 2004;117:2173–2181. doi: 10.1242/jcs.01227. [DOI] [PubMed] [Google Scholar]
- 31.Harbour J. W., Dean D. C. The Rb/E2F pathway: expanding roles and emerging paradigms. Genes Dev. 2000;14:2393–2409. doi: 10.1101/gad.813200. [DOI] [PubMed] [Google Scholar]
- 32.Roland B. D., Bernards R. Re-evaluating cell cycle regulation by E2Fs. Cell. 2006;127:1–4. doi: 10.1016/j.cell.2006.11.019. [DOI] [PubMed] [Google Scholar]
- 33.Narita M., Nunez S., heard E., Narita M., Lin A. W., Hearn S. A., Spector D. L., Hannon G. J., Lowe S. W. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell. 2003;113:703–716. doi: 10.1016/s0092-8674(03)00401-x. [DOI] [PubMed] [Google Scholar]
- 34.Zhang H. S., Postigo A. P., Dean D. C. Active transcriptional repression by the Rb–E2F complex mediates G1 arrest triggered by p16INK4a, TGF-β, and contact inhibition. Cell. 1999;97:53–61. doi: 10.1016/s0092-8674(00)80714-x. [DOI] [PubMed] [Google Scholar]
- 35.Lowe S. W., Sherr C. J. Tumor suppression by INK4a-Arf: progress and puzzles. Curr. Opin. Genet. Dev. 2003;13:77–83. doi: 10.1016/s0959-437x(02)00013-8. [DOI] [PubMed] [Google Scholar]
- 36.Young A. P., Nagarajan R., Longmore G. D. Mechanism of transcriptional regulation by Rb–E2F segregates by biological pathway. Oncogene. 2003;22:7209–7217. doi: 10.1038/sj.onc.1206804. [DOI] [PubMed] [Google Scholar]
- 37.Sharpless N. E., DePinho R. A. Telomeres, stem cells, senescence and cancer. J. Clin. Invest. 2004;113:160–168. doi: 10.1172/JCI20761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sage J., Mulligan G. J., Attardi L. D., Miller A., Chen S., Williams B., Theodorou E., Jacks T. Targeted disruption of the three Rb-related genes leads to loss of G(1) control and immortalization. Genes Dev. 2000;14:3037–3050. doi: 10.1101/gad.843200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bieda M., Xu X., Singer M. A., Green R., Farnham P. J. Unbiased location analysis of E2F1-binding sites suggests a widespread role for E2F1 in the human genome. Genome Res. 2006;16:595–605. doi: 10.1101/gr.4887606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trimarchi J. M., Lees J. A. Sibling rivalry in the E2F family. Nat. Rev. Mol. Cell Biol. 2002;3:11–20. doi: 10.1038/nrm714. [DOI] [PubMed] [Google Scholar]
- 41.Balciunaite E., Spektor A., Lents N. H., Cam H., te Riele H., Scime A., Rudnicki M. A., Young R., Dynlacht B. D. Pocket protein complexes are recruited to distinct target sites in quiescent and proliferating cells. Mol. Cell Biol. 2005;25:8166–817. doi: 10.1128/MCB.25.18.8166-8178.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morrison A. J., Sardet C., Herrera R. E. Retinoblastoma protein transcriptional repression through histone deacetylation of a single nucleosome. Mol. Cell. Biol. 2002;22:856–865. doi: 10.1128/MCB.22.3.856-865.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Janzen V., Forkert R., Fleming H. E., Saito Y., Waring M. T., Dombkowski D. M., Cheng T., DePinho R. A., Sharpless N. E., Scadden D. T. Stem-cell aging modified by the cyclin-dependent kinase inhibitor p16INK4a. Nature. 2006;443:421–426. doi: 10.1038/nature05159. [DOI] [PubMed] [Google Scholar]
- 44.Krishnamurthy J., Ramsey M. R., Ligon K. L., Torrice C., Koh A., Bonner-Weir S., Sharpless N. E. p16INK4a induces an age-dependent decline in islet regenerative potential. Nature. 2006;443:453–457. doi: 10.1038/nature05092. [DOI] [PubMed] [Google Scholar]
- 45.Molofsky A. V., Slutsky S. G., Joseph M. M., He S., Pardal R., Krishnamurthy J., Sharpless N. E., Morrison S. J. Increasing p16INK4a expression decreases forebrain progenitors and neurogenesis during ageing. Nature. 2006;443:448–452. doi: 10.1038/nature05091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Costantino M. E., Stearman R. P., Smith G. E., Darling D. S. Cell-specific phosphorylation of Zfhep transcription factor. Biochem. Biophys. Res. Comm. 2002;296:368–373. doi: 10.1016/s0006-291x(02)00880-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.