Abstract
Eukaryotic signaling and trafficking proteins are rich in modular domains that bind cell membranes. These binding events are tightly regulated in space and time. The structural, biochemical, and biophysical mechanisms for targeting have been worked out for many families of membrane binding domains. This review takes a comparative view of seven major classes of membrane binding domains, the C1, C2, PH, FYVE, PX, ENTH, and BAR domains. These domains use a combination of specific headgroup interactions, hydrophobic membrane penetration, electrostatic surface interactions, and shape complementarity to bind to specific subcellular membranes.
Keywords: Protein kinase C, diacylglycerol, phosphoinositide, signal transduction, membrane trafficking
1. Introduction
Eukaryotic cells are defined by their membrane-delimited organelles. Signals are transduced across membranes by machinery that must be at the membrane. Organelle biogenesis and identity depends on the protein and lipid composition of organellar membranes. Most eukaryotic proteins are modular in structure and contain one or more conserved domains. Given the diversity of membrane structures in eukaryotic cells and the complexity of domain structures in eukaryotic proteins, it makes sense that there so are many conserved membrane and lipid binding domains in the eukaryotic proteome. These domains are most abundant in proteins of signal transduction and protein and membrane trafficking, since these are processes that are carried out at cell membranes. My aim in this chapter is to provide an overview of these domains. I will draw selectively on the literature for a subset of the best understood domains in order to illustrate principles, while the reader is referred to other chapters in this volume for comprehensive reviews of individual domains.
2. Archetypal lipid binding domains
The history of membrane-binding domain research began with Nishizuka’s discovery of protein kinase C (PKC) in the late 1970’s. This discovery was followed by the cloning of several PKC isozymes and the observation of two regions of conserved sequence in the regulatory part of the enzyme. One of these regions, denoted C1 (for conserved region 1) at the time, was found to be responsible for phorbol ester binding [1]. We now know that this conserved region corresponds to two structural C1 domains that can bind to phorbol esters, diacylglycerol (DAG), and membranes [2, 3]. The second region, C2, is responsible for Ca2+ -dependence of conventional PKC activation [1]. This region corresponds directly to the C2 structural domain that binds to Ca2+ in isolation, as shown first for another C2 domain protein, synaptotagmin I [4]. Just as the discovery of PKC launched the field of lipid second messenger signaling, the discovery of the C1 and C2 domains launched the field of membrane binding domains in signal transduction. It is fitting that the third major membrane binding domain to be described, the pleckstrin homology (PH) domain, also owes its discovery in part to the PKC field. Pleckstrin was characterized as the major PKC substrate in platelets in the late 1980s, and the PH domain was discovered and named as a conserved region present in pleckstrin and other proteins implicated in signaling [5, 6]. The PH domain of phospholipase C-δ was the first to be established as a high-affinity membrane binding domain [7-10], in this case one specific for the lipid phosphatidylinositol (4,5)-bisphosphate (PI(4,5)P2). The year 1995 saw the first structural studies of high-affinity complexes of the C1 [11], C2 [12], and PH domains [13] with their ligands phorbol ester, Ca2+, and inositol (1,4,5)-trisphosphate (the headgroup of PI(4,5)P2). These and other studies on C1, C2, and PH domains in the years 1989-1995 spawned the membrane binding domain field and sketched its broad biochemical and biophysical principles.
The past decade has seen the expansion of membrane-binding domain studies in vivo and on a genomic scale, a continuing increase in the sophistication of biochemical and biophysical studies, and the introduction of many new members to this class of domains. 1998 was a seminal year in the membrane-binding domain field in that the use of green fluoresecent protein (GFP) fusions was introduced into the field and first used to probe the dynamics of diacylglycerol [14], Ca2+ [14], and PI(4,5)P2 [15, 16] in real time in living cells (see Balla chapter). A host of new membrane binding domains have been identified, including FERM (Four point one-ezrin-radixin-moesin), PX (phox), FYVE (Fab1/YOTB/Vac1/EEA1), Tubby, BAR (Bin/Amphiphysin/Rvs), and ENTH/ANTH (Epsin/AP180 N-terminal homology) [17-21]. The functions of the PH and PX domain families have been analyzed across the yeast genome, putting these domains into genomic perspective [22].
The major dilemma for structural biologists and biophysicists working in this field is that the structures of domains embedded in membranes are not approachable by x-ray crystallography. The smallest domains are approachable by solution NMR when embedded in short-chain lipid micelles, although not in bilayer membranes. This has been a major handicap to the advance of the field compared to areas of structural biology where functional complexes can be visualized at high resolution. A number of creative approaches are now being applied to fill in the gaps. The application of site-specific labeling for fluorescence resonance energy transfer (FRET) and electron paramagnetic resonance (EPR; [23] and see Cafiso chapter) was pioneered in studies of C2 domain:membrane interactions. These techniques now appear to be mature enough to be applied to other membrane-binding domains and perhaps to larger membrane-associated assemblies nucleated by these domains. Computational analysis of electrostatic interactions is rapidly becoming a standard part of membrane:domain interaction studies (see Murray chapter). All-atom molecular dynamics of membrane binding domains embedded in phospholipid bilayers is computationally demanding, but with current computational capabilities is now beginning to contribute to the field [24]. The application of x-ray reflectivity studies to membrane-bound domains, again using the C2 domain as a test case [25], is in its early stages but appears to be a very promising complement to these other approaches.
2.1 C1 domains
The C1 zinc finger domains of the conventional and novel (i.e. α, βI, βII, γ, δ, ε, θ, η, and μ, but not λ/ι or ζ) PKCs, chimaerins, RasGRPs and Unc-13 are known as the “typical” C1 domains, and are specific receptors for DAG and phorbol esters (see [26] and Kazanietz chapter). These C1 domains bind stereospecifically to phorbol esters, and presumably DAG, through a narrow polar groove between two pulled-apart β-strands [11]. The groove is surrounded by an exposed ridge of hydrophobic residues [11] that penetrate into the hydrocarbon core of the membrane [27, 28] (Figure 1). The Zn-finger structure of the C1 domain is thought to stabilize its fold in both its inactive, soluble state and its active, deeply membrane-embedded state. A belt of basic residues covers the middle portion of the domain, adjacent to the hydrophobic ridge at tip. These residues are thought to bind to the acidic headgroups of phosphatidylserine [29] at the surface of the membrane. C1 domains bind to phorbol esters embedded in acidic phospholipid membranes or micelles with ~104-fold higher affinity than to short-chain phorbol esters in free solution [30]. The interplay between the polar groove, hydrophobic ridge, and basic belt of the C1 domain is essential for high affinity binding to membrane-embedded phorbol esters and DAG.
Figure 1. Molecular surface models of membrane binding domains.
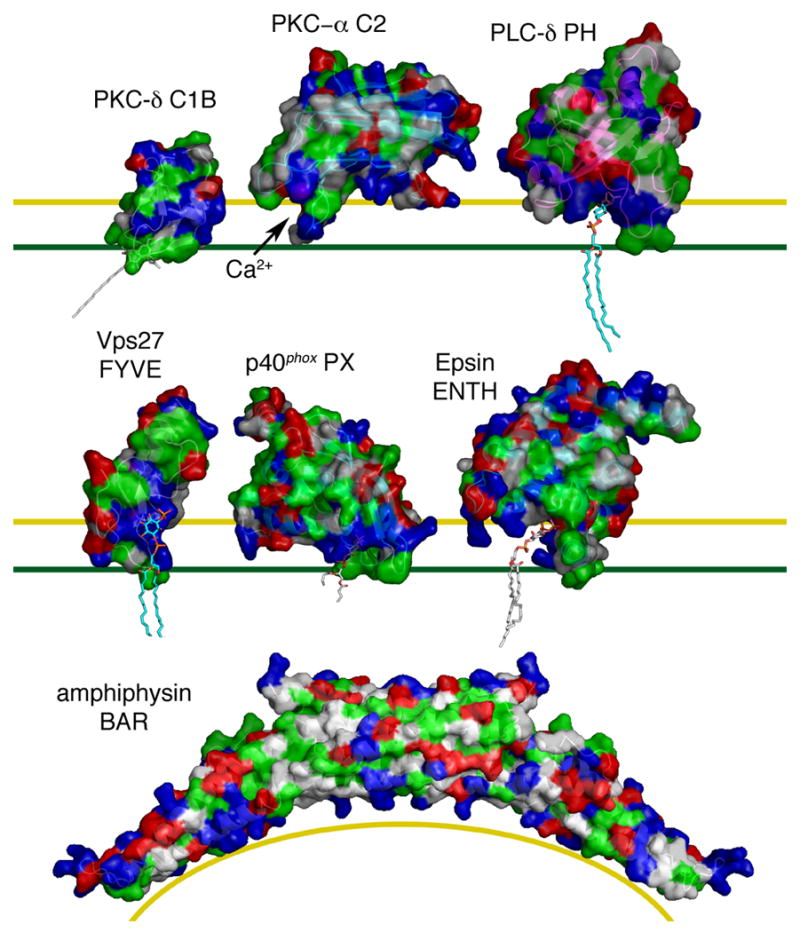
Surfaces are rendered with hydrophobic residues in green, basic in blue, acidic in red, and uncharged polar in white. The yellow line marks the top of the polar interface region of the bilayer, and the green line marks the top of the hydrocarbon core of the bilayer. The C1B domain of PKC-δ bound to phorbol 13-acetate (1PTR) [11] was docked to the membrane on the basis of structural modeling and solution NMR of the related PKC-γ C1B domain [27]. The C2 domain of PKC-α (1DSY) [39] was docked on the basis of FRET and EPR analysis [78]. The PH domain of PLC-δ (1MAI) [13] was docked on the basis of structural modeling. The FYVE domain of Vps27 (1VFY) [56], with PI(3)P docked on the basis of its binding site in EEA1 [58], was docked to the membrane based on structural and electrostatic modeling [74]. The PX domain of p40phox bound to PI(3)P (1H6H) [62] was docked on the basis of structural modeling and demonstrated membrane penetration [77]. The ENTH domain of epsin (1HOA) [63] bound to PI(4,5)P2 was docked on the basis of structural modeling and demonstrated membrane penetration [79]. The amphiphysin BAR domain (1URU) [68] was docked to a curved membrane surface based on shape complementarity. Phospholipid and phorbol ester tail moieties were modeled arbitrarily except for the PX domain, where the crystallized conformation is shown.
The C1 domain provided an early and vivid example of how the three major structural features of membrane binding domains work together: (1) a polar pocket or groove that stereospecifically recognizes a specific ligand; (2) a hydrophobic protrusion that penetrates into the hydrocarbon core of the membrane; and (3) clusters of basic residues that bind to acidic phospholipid headgroups (Figure 2). Of course, not all of these features are present in all membrane-binding domains. These features and the mechanisms associated with them are used to widely varying degrees in different families of membrane binding domains, and even among different instances of a given type of domain.
Figure 2. Conceptual model for membrane binding domain interactions.
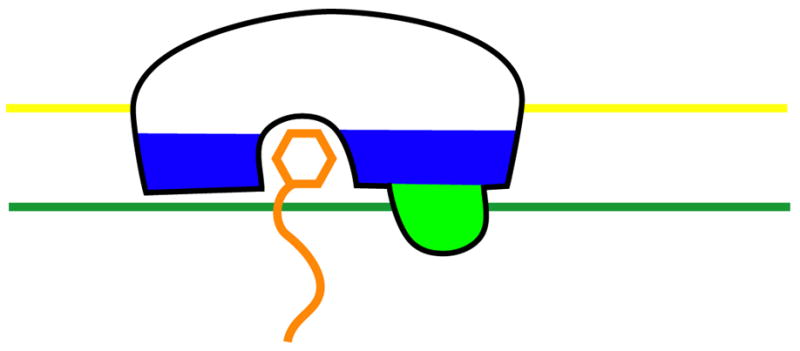
An idealized membrane binding domain is shown in the same color scheme used in Fig. 1.
The earliest studies of PKC focused on its activation by lipids and Ca2+, as opposed to subcellular targeting. The membrane engagement of the C1 domain was long known to allosterically activate the kinase, and the general outlines of a biochemical activation mechanism are known [31, 32]. The detailed structural mechanisms for enzyme activation by C1 and other membrane binding domains have lagged far behind analysis of targeting mechanisms, however. The mechanism for C1 domain-based activation has been worked out for one case, the RacGAP protein β2-chimaerin [33]. In the soluble and enzymatically inactive conformation, the β2-chimaerin C1 domain cooperates with an N-terminal inhibitory region to sterically block Rac binding. In this conformation, the hydrophobic ridge of the C1 engages in intra-protein interactions with other domains of β2-chimaerin. Membrane binding requires that these inhibitory intraprotein interactions be broken up, allowing Rac access to the catalytic RacGAP domain. The structural mechanism for PKC activation is likely to be similar, but elucidating the details remains an important challenge for the field.
2.2 C2 domains
The C2 domain is best known as the Ca2+ sensor in the conventional PKCs (α, βI, βII, γ) and synaptotagmin I, although many C2 domains in other proteins do not bind Ca2+ at all (see [34] and Cho & Stahelin chapter). C2 domains are β-sandwiches whose tip binds to between 2-3 Ca2+ ions via three loops known as Ca2+ binding regions (CBRs) [12, 35, 36]. Most Ca2+-binding C2 domains, the binding of the Ca2+ ions potentiates binding to acidic phospholipid membranes, with typically low specificity for one acidic lipid over another [37]. The C2 domain of cytosolic phospholipase A2 (cPLA2), however, binds preferentially to neutral membranes in its Ca2+-bound state [37]. The C2 domains of synaptotagmin I, PKCα, and PKCβ have basic residues surrounding the CBRs [12, 38, 39], while the cPLA2 has a unique amphipathic helix whose hydrophobic face penetrates the membrane in the Ca2+-bound state [40, 41]. A number of mechanisms have been proposed to explain how Ca2+ can promote binding to acidic membranes in some cases, and neutral membranes in others, including direct Ca2+-phospholipid bridging, Ca2+-induced conformational changes, and electrostatic control (reviewed in [17]). A computational electrostatic model has been developed that appears to explain the observed specificity differences through changes both in interaction energies and in the penalty for desolvating charges near the membrane [42]. Considerable effort has gone into defining the geometry of C2 domain interactions with membranes, making this field a test bed for applying FRET, EPR, x-ray reflectivity, and computational methods ([23]; Cafiso chapter).
2.3 PH domains
The PH domain family is the largest and most functionally diverse in lipid signaling (see [20, 43] and Lambright chapter). The PH domain fold also crops up as a substructure within the FERM domain [44]. One of the first functions reported for a PH domain was for that of the kinase GRK2 (formerly βARK1), which binds to the Gβγ complex through its PH domain [45]. NMR studies of the pleckstrin PH domain highlighted its ability to bind PI(4,5)P2 in micelles with moderate affinity and specificity [46]. A subset of PH domains, such as those from PLC-δ, GRP1, BTK, and PDK1, bind phosphoinositides with high affinity and specificity, and have perhaps received the most attention [19, 20]. It is ironic that the high-affinity group is a small fraction of the total, and that the great majority of PH domains either bind phosphoinositides with low to moderate affinity and specificity, or do not bind phosphoinositides at all [47].
The specificity determinants of these high affinity PH domains reside in basic residues in loops connecting the β-strands, which surround a deep pocket (for examples see [48, 49]). The surface corresponding to this pocket is less defined in the lower specificity PH domains, such as pleckstrin and spectrin [46, 50, 51]. The protein:protein interaction surface on the GRK2 PH domain is distinct from its membrane-binding surface [52], such that both interactions can occur simultaneously and synergistically. Unlike C1 domains, PH domains do not typically have large hydrophobic protrusions. This explains why PH domains such as that of PLC-δ bind to soluble lipid headgroups and membranes with comparable affinities [9], in contrast to the 104-fold difference for C1 domains.
3. Membrane binding domains in subcellular trafficking
The roots of the membrane binding domain field are in classical receptor-mediated signal transduction at the plasma membrane. Much of the growth in the field has centered on the role of membrane binding domains in vesicular trafficking, however. Some of the highlights in this area have been the discovery and structural characterization of FYVE [53-58] and PX [59-62] domains as receptors for the endosomal lipid phosphatidylinositol 3-phosphate; the characterization of high-affinity PI(4,5)P2 binding sites on endocytic proteins and complexes such as epsin [63] and AP180 [64-66], and the role of BAR domains and amphipathic α-helices in sensing and inducing membrane curvature [63, 67-70].
3.1 FYVE domains
FYVE domains mediate the endosomal targeting of diverse proteins by binding with high affinity and specificity to PI(3)P ([71] and Kutateladze chapter). FYVE domains are Zn fingers that are distant structural cousins of C1 domains [56]. The FYVE domain has a shallow basic pocket that specifically recognizes PI(3)P, additional basic residues for non-specific interactions with acidic phospholipid headgroups, and a hydrophobic protrusion that penetrates into the membrane[56-58]. The hydrophobic “turret loop” is located at essentially the same position as the membrane penetrating ridge in the C1 domain, although it is less extensive. As for the C1 domain, binding to PI(3)P and bulk phospholipid is synergistic [72] and involves membrane penetration [73]. The docking of FYVE domains onto short-chain lipid micelles has been extensively studied by NMR (Kutateladze chapter), since this domain is only ~60 amino acids and thus amenable to solution NMR approaches even with the added mass of a small lipid micelle. The role of electrostatics in FYVE domain:membrane binding has also been extensively probed by computational analysis ([74] and Murray chapter). The charge distribution on the surface of the FYVE domain appears to control the angle at which it docks onto membranes. While most FYVE domains bind specifically to PI(3)P, their membrane docking behavior seems to vary considerably. Figure 1 shows the perpendicular orientation of the Vps27 FYVE domain. In contrast, the FYVE domain of EEA1 is tilted by about 50° to the perpendicular when it binds membranes [58, 74, 75]. Despite differences in docking angle, all FYVE domains analyzed use their turret loops to penetrate the membrane.
3.2 PX domains
Like FYVE domains, the PX domain family is specialized for endosomal targeting and phosphoinositide binding (Hong chapter). Yeast PX domains all bind PI(3)P [76], but mammalian PX domains have a more diverse repertoire of ligands. The PX domain is about twice the size of the FYVE domain and has a completely different fold, showing how convergent evolution has led to two potent but completely different mechanisms for binding PI(3)P [62]. PX domains have a deep pocket for specific binding to phosphoinositides, a basic surface surrounding it, and a significant hydrophobic protrusion near the pocket that penetrates membranes [77].
3.3 ENTH domains
ENTH domains are a motif of endocytic proteins that have a stable octahelical core and a conformationally dynamic N-terminal amphipathic helix (De Camilli & Itoh chapter). The core and the N-terminal helix fold together to bind to PI(4,5)P2 [63]. The PI(4,5)P2-bound conformation of the epsin ENTH domain penetrates deeply into membranes using the exposed hydrophobic face of the amphipathic helix [63]. This membrane penetration is thought to promote the positive curvature of nascent vesicles in endocytosis. Like FYVE domains, the ENTH contains both a specific phosphoinositide headgroup-binding pocket composed of basic residues and a hydrophobic membrane-penetrating protrusion. The distinctive feature of the ENTH domain compared to the FYVE domain and others is that the hydrophobic protrusion is conformationally labile and only becomes structured only upon phosphoinositide binding.
3.4 BAR domains
BAR domains are helical dimers that do not recognize specific membrane lipids, but rather sense the curvature of a membrane (De Camilli & Itoh chapter and [69]). BAR dimers are shaped liked bananas and bind curved vesicular membranes with their concave faces [68, 69]. Portions of the concave faces are basic such that they can bind the surface of acidic phospholipid membranes. Thus the BAR domain illustrates the use of basic surfaces for non-specific acidic membrane binding, as seen in many other domains. The BAR domains are specialized such that these surfaces complement those of curved tubules, the necks of budding vesicles, or other highly curved membranes [68]. While BAR domains lack specific lipid recognition elements, they often occur in conjunction with PX or other domains to provide targeting specificity. Isolated BAR domains appear to be curvature sensors, rather than inducers, but BAR domains coupled to membrane-penetrating amphipathic helices are potent inducers of curvature.
4. Structural principles for membrane binding domains
The comparative analysis of different membrane binding domains reveals several persistent themes. The C1, PH, FYVE, PX, and ENTH domains are examples of domains that contain a pocket for the stereospecific recognition of a unique lipid ligand. These domains also recognize the soluble headgroups of their lipid ligands. In some cases, where additional contacts with the membrane are modest, as for the PLC-δ PH domain, the soluble headgroup binds with comparable affinity to the membrane-bound lipid. In other cases, the presence of the surrounding membrane is required to make additional contacts. The typical C1 domains, the FYVE and PX domains, and the epsin ENTH domain are examples of domains with hydrophobic protrusions that penetrate substantially into the hydrocarbon core of the membrane. Membrane penetration can provide very large enhancements to the affinity of domain:membrane interaction, as highlighted by the 104-fold preference for membrane-embedded vs. short-chain phorbol esters by the PKC-δ C1B domain. Membrane penetration can modulate membrane structure in biologically important ways, as first shown for the epsin ENTH domain in endocytic pit formation. The PKC-α C2 domain and the amphiphysin BAR domain are examples of membrane binding domains that rely primarily on interactions with the electrostatic field created by multiple acidic phospholipid headgroups. For the C2 domain, Ca2+ appears to function as a switch to regulate binding to planar membranes, whereas the BAR domain is shaped such that it can only bind to highly curved membranes.
Since the dawn of the membrane binding domain field in 1989, there has been enormous progress in understanding the biological, biochemical, and biophysical mechanisms of these domains. The contributors to this volume and many other colleagues deserve congratulations for having taken the field so far. While most of the broad principles appear to be in hand, nature has a way of surprising us just when we think we have it all figured out. After all, it has only been two years since the explanation for curvature sensing by BAR domains was worked out. In my view, the major frontier in the field now is to integrate the wealth of information we already have on individual membrane binding domains. We need to understand how membrane binding domains function in the context of large multiprotein assemblies on membranes, and how they work in the context of the large multidomain proteins that do the work in eukaryotic signaling and trafficking.
Acknowledgments
I thank the past and present members of my laboratory who have contributed to membrane-binding domain research, and Sangho Lee for comments on the manuscript. Research in my laboratory is supported by the intramural program of the NIDDK, NIH and by the IATAP program, NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ono Y, Fujii T, Igarashi K, Kuno T, Tanaka C, Kikkawa U, Nishizuka Y. Phorbol ester binding to protein kinase-C requires a cysteine-rich zinc-finger-like sequence. Proc Natl Acad Sci U S A. 1989;86:4868–4871. doi: 10.1073/pnas.86.13.4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Quest AFG, Bardes ESG, Bell RM. A phorbol ester binding domain of protein kinase-C-γ-high-affinity binding to a glutathione-S-transferase/cys2 fusion protein. J Biol Chem. 1994;269:2953–2960. [PubMed] [Google Scholar]
- 3.Kazanietz MG, Bustelo XR, Barbacid M, Kolch W, Mischak H, Wong G, Pettit GR, Bruns JD, Blumberg PM. Zinc-finger domains and phorbol ester pharmacophore - analysis of binding to mutated form of protein kinase-c-τ and the vav and c-raf protooncogene products. J Biol Chem. 1994;269:11590–11594. [PubMed] [Google Scholar]
- 4.Davletov BA, Sudhof TC. A single C2 domain from synaptotagmin-I is sufficient for high-affinity Ca2+/phospholipid binding. J Biol Chem. 1993;268:26386–26390. [PubMed] [Google Scholar]
- 5.Haslam RJ, Koide HB, Hemmings BA. Pleckstrin domain homology. Nature. 1993;363:309–310. doi: 10.1038/363309b0. [DOI] [PubMed] [Google Scholar]
- 6.Mayer BJ, Ren RB, Clark KL, Baltimore D. A putative modular domain present in diverse signaling proteins. Cell. 1993;73:629–630. doi: 10.1016/0092-8674(93)90244-k. [DOI] [PubMed] [Google Scholar]
- 7.Cifuentes ME, Honkanen L, Rebecchi MJ. Proteolytic fragments of phosphoinositide-specific phospholipase c-δ-1 - catalytic and membrane-binding properties. J Biol Chem. 1993;268:11586–11593. [PubMed] [Google Scholar]
- 8.Garcia P, Gupta R, Shah S, Morris AJ, Rudge SA, Scarlata S, Petrova V, McLaughlin S, Rebecchi MJ. The pleckstrin homology domain of phospholipase c-δ (1) binds with high affinity to phosphatidylinositol 4,5-bisphosphate in bilayer membranes. Biochemistry. 1995;34:16228–16234. doi: 10.1021/bi00049a039. [DOI] [PubMed] [Google Scholar]
- 9.Lemmon MA, Ferguson KM, Obrien R, Sigler PB, Schlessinger J. Specific and high-affinity binding of inositol phosphates to an isolated pleckstrin homology domain. Proc Natl Acad Sci U S A. 1995;92:10472–10476. doi: 10.1073/pnas.92.23.10472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yagisawa H, Hirata M, Kanematsu T, Watanabe Y, Ozaki S, Sakuma K, Tanaka H, Yabuta N, Kamata H, Hirata H, Nojima H. Expression and characterization of an inositol 1,4,5-trisphosphate binding domain of phosphatidylinositol-specific phospholipase c-δ (1) J Biol Chem. 1994;269:20179–20188. [PubMed] [Google Scholar]
- 11.Zhang GG, Kazanietz MG, Blumberg PM, Hurley JH. Crystal-structure of the cys2 activator-binding domain of protein-kinase c-δ in complex with phorbol ester. Cell. 1995;81:917–924. doi: 10.1016/0092-8674(95)90011-x. [DOI] [PubMed] [Google Scholar]
- 12.Sutton RB, Davletov BA, Berghuis AM, Sudhof TC, Sprang SR. Structure of the first C2 domain of synaptotagmin-I. A novel Ca2+/phospholipid-binding fold. Cell. 1995;80:929–938. doi: 10.1016/0092-8674(95)90296-1. [DOI] [PubMed] [Google Scholar]
- 13.Ferguson KM, Lemmon MA, Schlessinger J, Sigler PB. Structure of the high-affinity complex of inositol trisphosphate with a phospholipase-c pleckstrin homology domain. Cell. 1995;83:1037–1046. doi: 10.1016/0092-8674(95)90219-8. [DOI] [PubMed] [Google Scholar]
- 14.Oancea E, Meyer T. Protein kinase C as a molecular machine for decoding calcium and diacylglycerol signals. Cell. 1998;95:307–318. doi: 10.1016/s0092-8674(00)81763-8. [DOI] [PubMed] [Google Scholar]
- 15.Varnai P, Balla T. Visualization of phosphoinositides that bind pleckstrin homology domains: Calcium- and agonist-induced dynamic changes and relationship to myo-[H3]inositol-labeled phosphoinositide pools. J Cell Biol. 1998;143:501–510. doi: 10.1083/jcb.143.2.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stauffer TP, Ahn S, Meyer T. Receptor-induced transient reduction in plasma membrane PtdIns(4,5)P2 concentration monitored in living cells. Curr Biol. 1998;8:343–346. doi: 10.1016/s0960-9822(98)70135-6. [DOI] [PubMed] [Google Scholar]
- 17.Hurley JH, Misra S. Signaling and subcellular targeting by membrane-binding domains. Annu Rev Biophys Biomolec Struct. 2000;29:49–79. doi: 10.1146/annurev.biophys.29.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hurley JH, Meyer T. Subcellular targeting by membrane lipids. Curr Opin Cell Biol. 2001;13:146–152. doi: 10.1016/s0955-0674(00)00191-5. [DOI] [PubMed] [Google Scholar]
- 19.DiNitto JP, Cronin TC, Lambright DG. Membrane recognition and targeting by lipid-binding domains. Science STKE. 2003;re16:1–15. doi: 10.1126/stke.2132003re16. [DOI] [PubMed] [Google Scholar]
- 20.Lemmon MA. Phosphoinositide recognition domains. Traffic. 2003;4:201–213. doi: 10.1034/j.1600-0854.2004.00071.x. [DOI] [PubMed] [Google Scholar]
- 21.Cho WH, Stahelin RV. Membrane-protein interactions in cell signaling and membrane trafficking. Annu Rev Biophys Biomolec Struct. 2005;34:119–151. doi: 10.1146/annurev.biophys.33.110502.133337. [DOI] [PubMed] [Google Scholar]
- 22.Yu JW, Lemmon MA. Genome-wide analysis of signaling domain function. Curr Opin Chem Biol. 2003;7:103–109. doi: 10.1016/s1367-5931(02)00008-x. [DOI] [PubMed] [Google Scholar]
- 23.Malmberg NJ, Falke JJ. Use of EPR power saturation toanalyze the membrane-docking geometries of peripheral proteins: A applications to C2 domains. Annu Rev Biophys Biomolec Struct. 2005;34:71–90. doi: 10.1146/annurev.biophys.34.040204.144534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hritz J, Ulicny J, Laaksonen A, Jancura D, Miskovsky P. Molecular interaction model for the C1b domain of protein kinase c-γ in the complex with its activator phorbol-12-myristate-13-acetate in water solution and lipid bilayer. Journal of Medicinal Chemistry. 2004;47:6547–6555. doi: 10.1021/jm049786s. [DOI] [PubMed] [Google Scholar]
- 25.Malkova S, Long F, Stahelin RV, Pingali SV, Murray D, Cho WH, Schlossman ML. X-ray reflectivity studies of cPLA2 -α C2 domains adsorbed onto langmuir monolayers of sopc. Biophys J. 2005;89:1861–1873. doi: 10.1529/biophysj.105.061515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hurley JH, Newton AC, Parker PJ, Blumberg PM, Nishizuka Y. Taxonomy and function of C1 protein kinase C homology domains. Protein Sci. 1997;6:477–480. doi: 10.1002/pro.5560060228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu RX, Pawelczyk T, Xia TH, Brown SC. Nmr structure of a protein kinase c-γ phorbol-binding domain and study of protein-lipid micelle interactions. Biochemistry. 1997;36:10709–10717. doi: 10.1021/bi970833a. [DOI] [PubMed] [Google Scholar]
- 28.Medkova M, Cho WH. Interplay of C1 and C2 domains of protein kinase C-α in its membrane binding and activation. J Biol Chem. 1999;274:19852–19861. doi: 10.1074/jbc.274.28.19852. [DOI] [PubMed] [Google Scholar]
- 29.Johnson JE, Giorgione J, Newton AC. The C1 and C2 domains of protein kinase C are independent membrane targeting modules, with specificity for phosphatidylserine conferred by the C1 domain. Biochemistry. 2000;39:11360–11369. doi: 10.1021/bi000902c. [DOI] [PubMed] [Google Scholar]
- 30.Kazanietz MG, Barchi JJ, Omichinski JG, Blumberg PM. Low-affinity finding of phorbol esters to protein-kinase-C and its recombinant cysteine-rich region in the absence of phospholipids. J Biol Chem. 1995;270:14679–14684. doi: 10.1074/jbc.270.24.14679. [DOI] [PubMed] [Google Scholar]
- 31.Orr JW, Newton AC. Intrapeptide regulation of protein-kinase-C. J Biol Chem. 1994;269:8383–8387. [PubMed] [Google Scholar]
- 32.Newton AC, Johnson JJ. Protein kinase C: A paradigm for regulation of protein function by two membrane-targeting modules. Biochim Biophys Acta-Rev Biomembr. 1998;1376:155–172. doi: 10.1016/s0304-4157(98)00003-3. [DOI] [PubMed] [Google Scholar]
- 33.Canagarajah B, Collucio Leskow F, Ho YSJ, Mischak H, Saidi L, Kazanietz MG, Hurley JH. Structural mechanism for lipid activation of the Rac-specific GAP, β2-chimaerin. Cell. 2004;119:407–418. doi: 10.1016/j.cell.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 34.Nalefski EA, Falke JJ. The C2 domain calcium-binding motif: Structural and functional diversity. Protein Sci. 1996;5:2375–2390. doi: 10.1002/pro.5560051201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grobler JA, Essen LO, Williams RL, Hurley JH. C2 domain conformational changes in phospholipase C-δ 1. Nat Struct Biol. 1996;3:788–795. doi: 10.1038/nsb0996-788. [DOI] [PubMed] [Google Scholar]
- 36.Essen LO, Perisic O, Lynch DE, Katan M, Williams RL. A ternary metal binding site in the C2 domain of phosphoinositide-specific phospholipase C-δ 1. Biochemistry. 1997;36:2753–62. doi: 10.1021/bi962466t. [DOI] [PubMed] [Google Scholar]
- 37.Nalefski EA, Wisner MA, Chen JZ, Sprang SR, Fukuda M, Mikoshiba K, Falke JJ. C2 domains from different Ca2+ signaling pathways display functional and mechanistic diversity. Biochemistry. 2001;40:3089–3100. doi: 10.1021/bi001968a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sutton RB, Sprang SR. Structure of the protein kinase C-β phospholipid-binding C2 domain complexed with Ca2+ Structure. 1998;6:1395–1405. doi: 10.1016/s0969-2126(98)00139-7. [DOI] [PubMed] [Google Scholar]
- 39.Verdaguer N, Corbalan-Garcia S, Ochoa WF, Fita I, Gomez-Fernandez JC. Ca2+ bridges the C2 membrane-binding domain of protein kinase C α directly to phosphatidylserine. Embo J. 1999;18:6329–6338. doi: 10.1093/emboj/18.22.6329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Perisic O, Fong S, Lynch DE, Bycroft M, Williams RL. Crystal structure of a calcium-phospholipid binding domain from cytosolic phospholipase A2. J Biol Chem. 1998;273:1596–1604. doi: 10.1074/jbc.273.3.1596. [DOI] [PubMed] [Google Scholar]
- 41.Dessen A, Tang J, Schmidt H, Stahl M, Clark JD, Seehra J, Somers WS. Crystal structure of human cytosolic phospholipase A2 reveals a novel topology and catalytic mechanism. Cell. 1999;97:349–360. doi: 10.1016/s0092-8674(00)80744-8. [DOI] [PubMed] [Google Scholar]
- 42.Murray D, Honig B. Electrostatic control of the membrane targeting of C2 domains. Mol Cell. 2002;9:145–154. doi: 10.1016/s1097-2765(01)00426-9. [DOI] [PubMed] [Google Scholar]
- 43.Rebecchi MJ, Scarlata S. Pleckstrin homology domains: A common fold with diverse functions. Annu Rev Biophys Biomolec Struct. 1998;27:503-+. doi: 10.1146/annurev.biophys.27.1.503. [DOI] [PubMed] [Google Scholar]
- 44.Pearson MA, Reczek D, Bretscher A, Karplus PA. Structure of the erm protein moesin reveals the FERM domain fold masked by an extended actin binding tail domain. Cell. 2000;101:259–70. doi: 10.1016/s0092-8674(00)80836-3. [DOI] [PubMed] [Google Scholar]
- 45.Koch WJ, Inglese J, Stone WC, Lefkowitz RJ. The binding-site for the βγ-subunits of heterotrimeric γ-proteins on the β-adrenergic-receptor kinase. J Biol Chem. 1993;268:8256–8260. [PubMed] [Google Scholar]
- 46.Harlan JE, Hajduk PJ, Yoon HS, Fesik SW. Pleckstrin homology domains bind to phosphatidylinositol-4,5-bisphosphate. Nature. 1994;371:168–70. doi: 10.1038/371168a0. [DOI] [PubMed] [Google Scholar]
- 47.Yu JW, Mendrola JM, Audhya A, Singh S, Keleti D, DeWald DB, Murray D, Emr SD, Lemmon MA. Genome-wide analysis of membrane targeting by S. cerevisiae pleckstrin homology domains. Mol Cell. 2004;13:677–688. doi: 10.1016/s1097-2765(04)00083-8. [DOI] [PubMed] [Google Scholar]
- 48.Ferguson KM, Kavran JM, Sankaran VG, Fournier E, Isakoff SJ, Skolnik EY, Lemmon MA. Structural basis for discrimination of 3-phosphoinositides by pleckstrin homology domains. Mol Cell. 2000;6:373–84. doi: 10.1016/s1097-2765(00)00037-x. [DOI] [PubMed] [Google Scholar]
- 49.Lietzke SE, Bose S, Cronin T, Klarlund J, Chawla A, Czech MP, Lambright DG. Structural basis of 3-phosphoinositide recognition by pleckstrin homology domains. Mol Cell. 2000;6:385–394. doi: 10.1016/s1097-2765(00)00038-1. [DOI] [PubMed] [Google Scholar]
- 50.Ferguson KM, Lemmon MA, Schlessinger J, Sigler PB. Crystal structure at 2.2 Å resolution of the pleckstrin homology domain from human dynamin. Cell. 1994;79:199–209. doi: 10.1016/0092-8674(94)90190-2. [DOI] [PubMed] [Google Scholar]
- 51.Macias MJ, Musacchio A, Ponstingl H, Nilges M, Saraste M, Oschkinat H. Structure of the pleckstrin homology domain from β-spectrin. Nature. 1994;369:675–7. doi: 10.1038/369675a0. [DOI] [PubMed] [Google Scholar]
- 52.Lodowski DT, Pitcher JA, Capel WD, Lefkowitz RJ, Tesmer JJG. Keeping G proteins at bay: A complex between G protein-coupled receptor kinase 2 and Gβγ. Science. 2003;300:1256–1262. doi: 10.1126/science.1082348. [DOI] [PubMed] [Google Scholar]
- 53.Gaullier JM, Simonsen A, D'Arrigo A, Bremnes B, Stenmark H, Aasland R. FYVE fingers bind PtdIns(3)P. Nature. 1998;394:432–433. doi: 10.1038/28767. [DOI] [PubMed] [Google Scholar]
- 54.Patki V, Lawe DC, Corvera S, Virbasius JV, Chawla A. A functional ptdins(3)p-binding motif. Nature. 1998;394:433–434. doi: 10.1038/28771. [DOI] [PubMed] [Google Scholar]
- 55.Burd CG, Emr SD. Phosphatidylinositol(3)-phosphate signaling mediated by specific binding to RING FYVE domains. Mol Cell. 1998;2:157–162. doi: 10.1016/s1097-2765(00)80125-2. [DOI] [PubMed] [Google Scholar]
- 56.Misra S, Hurley JH. Crystal structure of a phosphatidylinositol 3-phosphate- specific membrane-targeting motif, the FYVE domain of Cps27p. Cell. 1999;97:657–666. doi: 10.1016/s0092-8674(00)80776-x. [DOI] [PubMed] [Google Scholar]
- 57.Kutateladze TG, Ogburn KD, Watson WT, de Beer T, Emr SD, Burd CG, Overduin M. Phosphatidylinositol 3-phosphate recognition by the FYVE domain. Mol Cell. 1999;3:805–811. doi: 10.1016/s1097-2765(01)80013-7. [DOI] [PubMed] [Google Scholar]
- 58.Dumas JJ, Merithew E, Sudharshan E, Rajamani D, Hayes S, Lawe D, Corvera S, Lambright DG. Multivalent endosome targeting by homodimeric EEA1. Mol Cell. 2001;8:947–958. doi: 10.1016/s1097-2765(01)00385-9. [DOI] [PubMed] [Google Scholar]
- 59.Kanai F, Liu H, Field SJ, Akbary H, Matsuo T, Brown GE, Cantley LC, Yaffe MB. The px domains of p47phox and p40phox bind to lipid products of PI(3)K. Nat Cell Biol. 2001;3:675–678. doi: 10.1038/35083070. [DOI] [PubMed] [Google Scholar]
- 60.Xu Y, Hortsman H, Seet L, Wong SH, Hong W. SNX3 regulates endosomal function through its PX domain-mediated interaction with PtdIns(3)P. Nat Cell Biol. 2001;3:658–66. doi: 10.1038/35083051. [DOI] [PubMed] [Google Scholar]
- 61.Ellson CD, Gobert-Gosse S, Anderson KE, Davidson K, Erdjument-Bromage H, Tempst P, Thuring JW, Cooper MA, Lim ZY, Holmes AB, Gaffney PRJ, Coadwell J, Chilvers ER, Hawkins PT, Stephens LR. PtdIns(3)P regulates the neutrophil oxidase complex by binding to the PX domain of p40phox. Nat Cell Biol. 2001;3:679–682. doi: 10.1038/35083076. [DOI] [PubMed] [Google Scholar]
- 62.Bravo J, Karathanassis D, Pacold CM, Pacold ME, Ellson CD, Anderson KE, Butler PJG, Lavenir I, Perisic O, Hawkins PT, Stephens L, Williams RL. The crystal structure of the PX domain from p40phox bound to phosphatidylinositol 3-phosphate. Mol Cell. 2001;8:829–839. doi: 10.1016/s1097-2765(01)00372-0. [DOI] [PubMed] [Google Scholar]
- 63.Ford MGJ, Mills IG, Peter BJ, Vallis Y, Praefcke GJK, Evans PR, McMahon HT. Curvature of clathrin-coated pits driven by epsin. Nature. 2002;419:361–366. doi: 10.1038/nature01020. [DOI] [PubMed] [Google Scholar]
- 64.Ford MGJ, Pearse BMF, Higgins MK, Vallis Y, Owen DJ, Gibson A, Hopkins CR, Evans PR, McMahon HT. Simultaneous binding of PtdIns(4,5)P2 and clathrin by AP180 in the nucleation of clathrin lattices on membranes. Science. 2001;291:1051–1055. doi: 10.1126/science.291.5506.1051. [DOI] [PubMed] [Google Scholar]
- 65.Mao YX, Chen J, Maynard JA, Zhang B, Quiocho FA. A novel all helix fold of the AP180 amino-terminal domain for phosphoinositide binding and clathrin assembly in synaptic vesicle endocytosis. Cell. 2001;104:433–440. doi: 10.1016/s0092-8674(01)00230-6. [DOI] [PubMed] [Google Scholar]
- 66.Itoh T, Koshiba S, Kigawa T, Kikuchi A, Yokoyama S, Takenawa T. Role of the ENTH domain in phosphatidylinositol-4,5-bisphosphate binding and endocytosis. Science. 2001;291:1047–1051. doi: 10.1126/science.291.5506.1047. [DOI] [PubMed] [Google Scholar]
- 67.Hurley JH, Wendland B. Endocytosis: Driving membranes around the bend. Cell. 2002;111:143–146. doi: 10.1016/s0092-8674(02)01044-9. [DOI] [PubMed] [Google Scholar]
- 68.Peter BJ, Kent HM, Mills IG, Vallis Y, Butler PJG, Evans PR, McMahon HT. Bar domains as sensors of membrane curvature: The amphiphysin bar structure. Science. 2004;303:495–499. doi: 10.1126/science.1092586. [DOI] [PubMed] [Google Scholar]
- 69.Gallop JL, McMahon HT. Lipids, rafts and traffic. 2005. pp. 223–231. [DOI] [PubMed] [Google Scholar]
- 70.Lee MCS, Orci L, Hamamoto S, Futai E, Ravazzola M, Schekman R. Sar1p N-terminal helix initiates membrane curvature and completes the fission of a COPII vesicle. Cell. 2005;122:605–617. doi: 10.1016/j.cell.2005.07.025. [DOI] [PubMed] [Google Scholar]
- 71.Misra S, Miller GJ, Hurley JH. Recognizing phosphatidylinositol 3-phosphate. Cell. 2001;107:559–562. doi: 10.1016/s0092-8674(01)00594-3. [DOI] [PubMed] [Google Scholar]
- 72.Sankaran VG, Klein DE, Sachdeva MM, Lemmon MA. High-affinity binding of a fyve domain to phosphatidylinositol 3-phosphate requires intact phospholipid but not FYVE domain oligomerization. Biochemistry. 2001;40:8581–7. doi: 10.1021/bi010425d. [DOI] [PubMed] [Google Scholar]
- 73.Stahelin RV, Long F, Diraviyam K, Bruzik KS, Murray D, Cho W. Phosphatidylinositol 3-phosphate induces the membrane penetration of the FYVE domains of Vps27p and Hrs. J Biol Chem. 2002;277:26379–88. doi: 10.1074/jbc.M201106200. [DOI] [PubMed] [Google Scholar]
- 74.Diraviyam K, Stahelin RV, Cho W, Murray D. Computer modeling of the membrane interaction of FYVE domains. J Mol Biol. 2003;328:721–736. doi: 10.1016/s0022-2836(03)00325-5. [DOI] [PubMed] [Google Scholar]
- 75.Brunecky R, Lee S, Rzepecki PW, Overduin M, Prestwich GD, Kutateladze AG, Kutateladze TG. Investigation of the binding geometry of a peripheral membrane protein. Biochemistry. 2005;44:16064–16071. doi: 10.1021/bi051127+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yu JW, Lemmon MA. All phox homology (PX) domains from Saccharomyces cerevisiae specifically recognize phosphatidylinositol 3-phosphate. J Biol Chem. 2001;276:44179–44184. doi: 10.1074/jbc.M108811200. [DOI] [PubMed] [Google Scholar]
- 77.Stahelin RV, Burian A, Bruzik KS, Murray D, Cho WW. Membrane binding mechanisms of the PX domains of nadph oxidase p40 phox and p47 phox. J Biol Chem. 2003;278:14469–14479. doi: 10.1074/jbc.M212579200. [DOI] [PubMed] [Google Scholar]
- 78.Kohout SC, Corbalan-Garcia S, Gomez-Fernandez JC, Falke JJ. C2 domain of protein kinase C α: Elucidation of the membrane docking surface by site-directed fluorescence and spin labeling. Biochemistry. 2003;42:1254–1265. doi: 10.1021/bi026596f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Stahelin RV, Long F, Peter BJ, Murray D, De Camilli P, McMahon HT, Cho W. Contrasting membrane interaction mechanisms of AP180 N-terminal homology (ANTH) and epsin N-terminal homology (ENTH) domains. J Biol Chem. 2003;278:28993–28999. doi: 10.1074/jbc.M302865200. [DOI] [PubMed] [Google Scholar]


