Abstract
The origin recognition complex (ORC) nucleates DNA replication initiation in eukaryotic cells. This six-protein complex binds replication origin DNA, recruits other initiation factors, and facilitates loading of the DNA helicase. Studying the function of individual ORC subunits during pre-RC formation has been hampered by the requirement of most subunits for DNA binding. In this study, we investigate the function of the Saccharomyces cerevisiae Orc6, the only ORC subunit not required for DNA binding. In vivo, depletion of Orc6 inhibits prereplicative complex (pre-RC) assembly and maintenance. In vitro, ORC lacking Orc6 fails to interact with Cdt1 and to load the Mcm2–7 helicase onto origin DNA. We demonstrate that two regions of Orc6 bind Cdt1 directly, and that the extreme C terminus of Orc6 (Orc6-CTD) interacts tightly with the remaining five ORC subunits. Replacing Orc6 with a fusion protein linking Cdt1 to the Orc6-CTD results in an ORC complex that loads Mcm2–7 onto DNA. Interestingly, this complex can only perform a single round of Mcm2–7 loading, suggesting that a dynamic association of Cdt1 with ORC is required for multiple rounds of Mcm2–7 loading.
[Keywords: ATPase, DNA replication, prereplicative complex (pre-RC), molecular machine, origin licensing]
Eukaryotic cells must coordinate the activity of numerous origins of replication to ensure that the genome is fully replicated exactly once per cell cycle. These sites are marked during late M and G1 cell cycle phases by the formation of a multiprotein complex called the prereplicative complex (pre-RC). As cells enter S phase, a subset of existing pre-RCs are activated to direct the formation of bidirectional replication forks while new pre-RC formation is inhibited. Therefore, only pre-RCs formed in G1 can subsequently direct replication initiation. Mutations that result in inadequate pre-RC formation lead to reduced replication forks and incomplete DNA replication, and are associated with genomic instability and cancer (Lengronne and Schwob 2002; Tanaka and Diffley 2002a; Ekholm-Reed et al. 2004; Shima et al. 2007).
Pre-RC formation requires the repeated loading of the Mcm2–7 helicase at each origin (Edwards et al. 2002; Bowers et al. 2004). The origin recognition complex (ORC) nucleates these events through its association both with origin DNA and with other replication proteins. As cells exit the M phase of the cell cycle, Cdc6 binds origin-bound ORC to form a coordinated Mcm2–7 loading machine (Bowers et al. 2004; Speck et al. 2005; Randell et al. 2006). Together, ORC and Cdc6 recruit Cdt1 and an initial Mcm2–7 complex to the origin. In an event requiring ordered ATP hydrolysis by Cdc6 and then ORC (Randell et al. 2006), the associated Mcm2–7 complex is loaded onto origin DNA (Donovan et al. 1997; Rowles et al. 1999; Bowers et al. 2004). Repeated cycles of hydrolysis result in multiple Mcm2–7 complexes being loaded per origin.
ORC performs multiple functions during pre-RC formation, recognizing origin DNA, recruiting other pre-RC proteins, and helping to load the Mcm2–7 complex. Nevertheless, the molecular interactions between the pre-RC components and ORC and how they contribute to helicase loading remain largely unknown. For example, it is clear that ORC is required to recruit Cdc6, Cdt1, and Mcm2–7 to the origin; however, only the interaction between ORC and Cdc6 is known to be direct (Mizushima et al. 2000; Speck et al. 2005). In addition to understanding the interactions between pre-RC proteins, it is also important to understand how these interactions are changed during the steps of pre-RC formation. For example, Cdt1 associates with the remaining pre-RC components only transiently (Gillespie et al. 2001; Randell et al. 2006). This association is controlled by the ATP-bound state of Cdc6, but the importance of Cdt1 release during pre-RC assembly has not been determined. It is likely that a number of other interactions in the pre-RC are similarly dynamic.
The smallest ORC subunit, Orc6, is distinct from the remaining ORC subunits. Orc6 is the only subunit that is not related to the AAA+ family of ATPases (Speck et al. 2005; Erzberger and Berger 2006). In Saccharomyces cerevisiae cells, Orc6 is also the only ORC subunit that is not required for ORC DNA binding (Lee and Bell 1997). Nevertheless, Orc6 is essential to cell division, and recent studies of yeast cells with reduced levels of Orc6 indicate that it has a role in maintaining Mcm2–7 association with chromatin (Semple et al. 2006; Da-Silva and Duncker 2007). Together these observations suggest that Orc6 functions downstream from ORC DNA binding, possibly by recruiting other pre-RC components. Orc6 takes on additional functions in other organisms. Studies of both the Drosophila and human Orc6 have identified a role for Orc6 in cytokinesis (Prasanth et al. 2002; Chesnokov et al. 2003). Unlike S. cerevisiae Orc6, its Drosophila counterpart plays a critical role in ORC DNA binding (Balasov et al. 2007). Despite this diversity of function across different species, in all organisms studied Orc6 is required for DNA replication, strongly suggesting that Orc6 function during DNA replication is conserved.
In this study, we investigate the mechanism of S. cerevisiae Orc6 function during pre-RC formation. Using genome-wide, in vivo analyses, we show that Orc6 is required for the assembly and maintenance of all pre-RCs. In vitro assays reveal that Orc6 is required for Cdt1 and Mcm2–7 but not Cdc6 association with origin DNA. We demonstrate that Orc6 directly interacts with Cdt1, Orc3, and Orc5. Tethering Cdt1 to the Orc1–5 bypasses the requirement for full-length Orc6 during in vitro Mcm2–7 loading. Analysis of the Cdt1–ORC fusion reveals important properties of pre-RC formation, including a requirement for Cdt1 release to enable repeated Mcm2–7 loading.
Results
An ORC6 degron allele arrests with unreplicated DNA
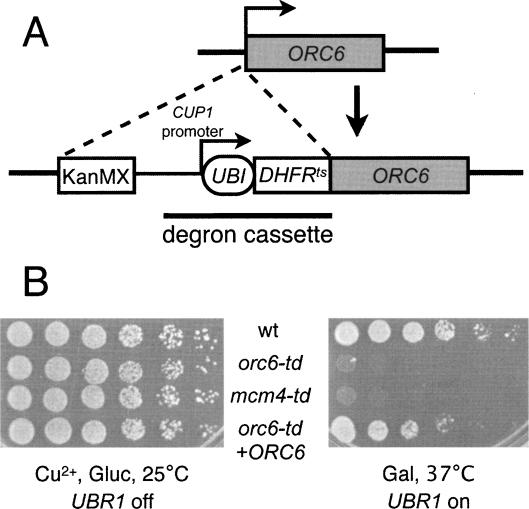
To study the role of Orc6 in cells, we made an ORC6 degron allele (orc6-td) that rapidly depletes Orc6 protein in galactose medium at elevated temperatures (Fig. 1A; see Materials and Methods). Consistent with the essential nature of Orc6, the Orc6 degron strain failed to form colonies under nonpermissive conditions (Fig. 1B) and 97% of the cells exhibited first cell cycle arrest in microcolony assays (Supplementary Table 1). A control Mcm4 degron strain (mcm4-td) showed similar defects under nonpermissive conditions. Addition of a plasmid expressing wild-type ORC6 restored the ability of orc6-td cells to grow.
Figure 1.
Inactivation of Orc6 causes growth defects. (A) The Orc6 temperature-sensitive degron strain was constructed by transforming wild-type cells with a PCR product containing KanMX, CUP1 promoter, and degron cassette flanked with Orc6 homologous sequences. The integration of the degron cassette was confirmed by PCR. (B) Serial dilutions (1:5) of a wild-type control strain, orc6-td, mcm4-td, and an orc6-td strain with a plasmid carrying wild-type ORC6 were grown under the indicated conditions and examined after 2 d.
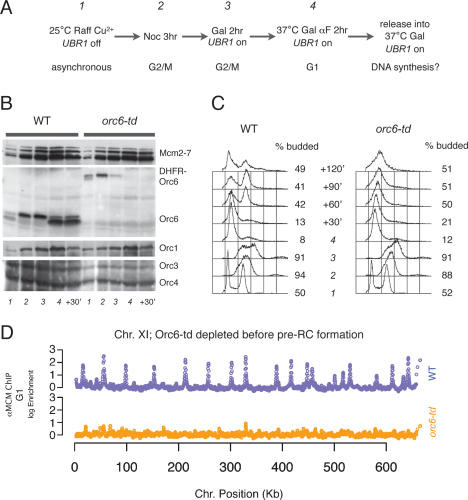
To determine when Orc6 was required during the cell cycle, we assessed the effect of the orc6-td allele on cell cycle progression. After arresting the cells at the G2/M boundary with nocodazole, degradation of Orc6 was induced and the cells were allowed to proceed into the following G1 in the presence of α factor (Fig. 2A). Cells were then released into S phase under nonpermissive conditions. Protein, FACS, and chromatin immunoprecipitation (ChIP) samples were taken at various times to monitor pre-RC protein levels, cell cycle progression, and chromatin association.
Figure 2.
Orc6 is required for pre-RC establishment. (A) Experimental outline. Wild-type and orc6-td cultures were grown to an OD600 = 0.6 at room temperature in YP + Raffinose containing Cu2+ (time point 1) before adding nocodazole to arrest cells at G2/M (time point 2). (Time point 3) Galactose was added to 2% to induce UBR1 expression for 2 h. (Time point 4) Cells were then released into fresh YP + Galactose in the presence of α factor at 37°C to fully deplete the Orc6-td protein. To assess whether cells could proceed to synthesize DNA, the cultures were released from α-factor arrest into fresh YP + Galactose at 37°C. (B) Orc6-td protein was depleted while other ORC subunits and Mcm2–7 remained. TCA-precipitated whole-protein samples from indicated time points were separated by SDS–polyacrylamide gel electrophoresis, blotted, and probed with monoclonal anti-Mcm2–7 (AS1.1), anti-Orc6 (SB49), anti-Orc1 (SB13), anti-Orc3 (SB3), and anti-Orc4 (SB6) antibodies. (C) The DNA content of samples from the indicated time points was analyzed by flow cytometry. Budding indices are shown to the right of each time point. (D) orc6-td cells failed to load Mcm2–7 on origins. Pre-RC formation was detected by anti-Mcm2–7 ChIP hybridized on yeast 44K tiled DNA microarray. Log2 ratio of enrichment of anti-Mcm2–7 ChIP sample over input from time point 4 in A (establishment) was plotted as a function of its position along the chromosome. Chromosome XI is shown here as an example (wild-type strain is blue, and orc6-td strain is orange).
Orc6-td protein was rapidly depleted upon shifting cells to nonpermissive conditions. In wild-type cultures, total Orc6 levels are unchanged during the cell cycle, although Orc6 migration changes between a phosphorylated (during S and G2/M) and dephosphorylated (during G1) form (Figs. 2B, 3B). In the orc6-td strain, Orc6-td appears as a DHFR-Orc6 fusion protein (after removal of N-terminal ubiquitin). Like Orc6, we observed two Orc6-td bands that show the same pattern of phosphorylation and dephosphorylation during the cell cycle (Figs. 2B, 3B). Importantly, the protein level of DHFR-Orc6 in asynchronous culture was comparable with that of the wild-type strain but decreased rapidly when degradation was induced. DHFR-Orc6 was undetectable in cells when the culture was raised to 37°C, and remained depleted at later time points (Fig. 2B). Depletion of Orc6 did not affect the levels of other pre-RC components as other ORC subunits, and the Mcm2–7 proteins were unchanged after Orc6 degradation was induced (Fig. 2B).
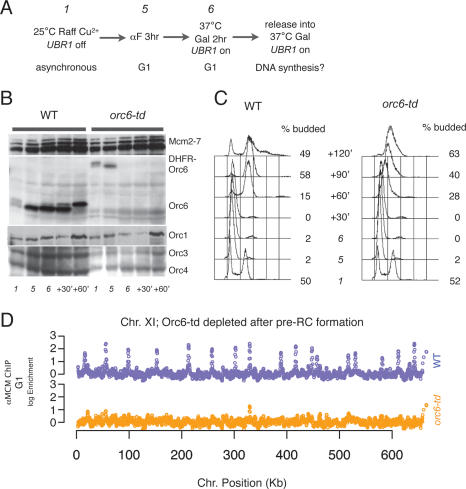
Figure 3.
Orc6 is required for pre-RC maintenance. (A) Experimental outline. Wild-type and orc6-td cultures were grown to an OD600 = 0.6 at room temperature in YP + Raffinose containing Cu2+ (time point 1) before adding α factor to arrest cells at G1 (time point 5). (Time point 6) Cells were switched into fresh YP + Galactose in the presence of α factor at 37°C to deplete Orc6-td. To assess whether cells could proceed to synthesize DNA, the cultures were released from α-factor arrest into fresh YP + Galactose at 37°C. (B) Orc6-td protein was depleted while other ORC subunits and Mcm2–7 remained. TCA-precipitated whole-protein samples from the indicated time points were assayed as described in Figure 2B. (C) The DNA content of samples from the indicated time points was analyzed by flow cytometry. Budding indices are shown to the right of each time point. (D) Depletion of Orc6 after pre-RC formation caused Mcm2–7 to dissociate from origins. Anti-Mcm2–7 ChIP samples and input from time point 6 in A were analyzed as described in Figure 2D. Chromosome XI is shown as an example (wild-type strain is blue, and orc6-td strain is orange).
Cells depleted of Orc6 showed defects in DNA replication. Passage from M phase into G1 was comparable for cells with and without Orc6, indicating that Orc6 does not play an essential role in cytokinesis in S. cerevisiae (Fig. 2C; data not shown). In contrast, when cells depleted of Orc6 were released from the G1 arrest, subsequent progression through S phase was dramatically impaired (Fig. 2C).
Orc6 is required for pre-RC assembly
To determine if the DNA replication defect in the Orc6-depleted cells was the result of impaired pre-RC formation, we monitored this event across the genome using Mcm2–7 ChIP. After shifting to nonpermissive conditions at G2/M, we isolated chromatin from wild-type and orc6-td cells at the G1 arrest point and precipitated these samples using anti-Mcm2–7 antibodies. The precipitated DNA was fluorescently labeled and hybridized to a genome-wide, tiled microarray along with a differentially labeled total genomic control DNA. As expected, in wild-type cells we observed robust association of Mcm2–7 at origins across the genome in G1 phase. In contrast, in orc6-td cells in which Orc6 had been depleted, Mcm2–7 association was reduced to background levels at all but a few origins (Fig. 2D). These findings demonstrate that pre-RC formation requires the Orc6 subunit, and suggest that defective pre-RC formation is the primary cause of the replication defect observed in the absence of Orc6.
To determine if the Orc6-depletion defect was due to a problem with ORC DNA binding or a downstream event, we also analyzed ORC DNA binding across the genome. We tested ORC association in wild-type cells at G2/M, Orc6-reduced condition at G2/M, and Orc6-depleted condition at G1 using anti-ORC ChIP (Supplementary Fig. 1). We observed the same pattern and extent of ORC binding in all three samples, indicating that, as is seen in vitro, Orc6 was not required for ORC DNA binding in vivo.
Orc6 is required for pre-RC maintenance
We next asked whether there was a role of Orc6 in maintaining pre-RC association with origins once they have formed. Previous studies of ORC1 and ORC2 mutant strains indicated that ORC is required for stable maintenance of pre-RCs (Aparicio et al. 1997). Because both of these mutants inhibit ORC DNA binding, we asked whether a mutation that did not displace ORC from the DNA (orc6-td) would have the same effect. Previous reports demonstrated that partial depletion of Orc6 reduced but did not eliminate Mcm2 association with chromatin (Semple et al. 2006; Da-Silva and Duncker 2007). Given this observation, we wondered whether any effects on pre-RC maintenance would be genome-wide or origin specific.
Unlike the above analysis in which we depleted Orc6 prior to pre-RC formation, in these experiments we arrested orc6-td and wild-type strains after pre-RC formation had occurred (in G1) and then induced Orc6 degradation. Pre-RC maintenance was evaluated by monitoring Mcm2–7 origin association across the genome and cell cycle progression was measured by FACS (Fig. 3). When Orc6 was depleted from G1-arrested cells, Mcm2–7 association was lost from all sites of pre-RC formation across the genome (Fig. 3D). No new sites of association were observed, indicating that the Mcm2–7 proteins were released from the DNA rather than moving to new sites away from the origin. Consistent with this loss of Mcm2–7 association, when released from G1 arrest, the orc6-td strain replicated its DNA poorly compared with the wild-type strain (Fig. 3C). Thus, Orc6 function is uniformly required to maintain functional pre-RCs at all origins.
ORCΔ6 is defective for pre-RC assembly in vitro
To determine the specific role of Orc6 during pre-RC formation, we used a partially reconstituted in vitro pre-RC assembly assay (Seki and Diffley 2000; Randell et al. 2006). This assay relies on purified ORC and Cdc6 as well as an extract derived from G1-arrested cells to assemble pre-RCs on immobilized DNA containing the ARS1 origin of replication. We purified ORC that was lacking the Orc6 subunit (ORCΔ6) from insect cells and the individual Orc6 subunit from Escherichia coli. In contrast to Drosophila Orc6, which binds DNA directly and independently (Balasov et al. 2007), yeast Orc6 showed no interactions with ARS1 DNA on its own (Supplementary Fig. 2). However, Orc6 did bind ARS1 DNA if purified ORCΔ6 was also present (Supplementary Fig. 2), suggesting that it could reassociate with the DNA-binding competent ORCΔ6.
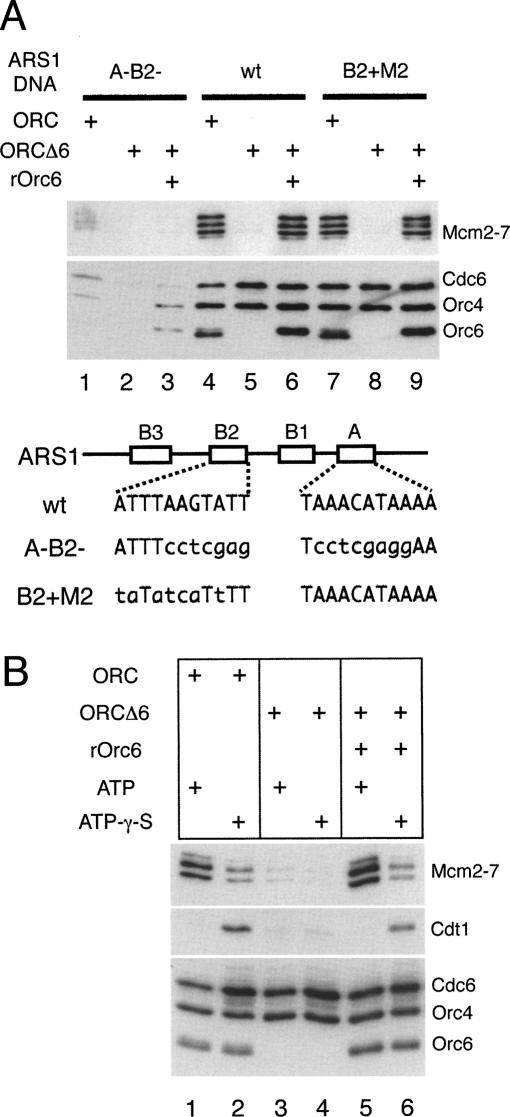
We tested the ability of purified ORCΔ6 to substitute for ORC in the pre-RC assembly assay. First, we depleted all ORC subunits from the pre-RC assembly extract (Bowers et al. 2004). When purified wild-type ORC was added to the assay, robust association of ORC, Cdc6, and Mcm2–7 proteins was observed (Fig. 4A, lane 4). When Orc6 was omitted from the complex (ORCΔ6), Orc4 and Cdc6 association with origin DNA were not affected (Fig. 4A, cf. lanes 4 and 5). In contrast, in the absence of Orc6 we observed no detectable Mcm2–7 binding, consistent with the pre-RC defect observed after Orc6 depletion in vivo. This defect could be rescued by the addition of recombinant Orc6, indicating ORC suffered no irreversible damage in the absence of Orc6 (Fig. 4A, lane 6). Finally, all associations were dependent on the presence of the wild-type ARS1 ORC-binding site (the ARS consensus sequence, or ACS) (Fig. 4A, lanes 4–6 wild-type vs. lanes 1–3 A−B2−).
Figure 4.
Orc6 is required for pre-RC assembly in vitro. (A) Pre-RC assembly assays with ORC and ORCΔ6 were performed with ORC-depleted G1 whole-cell extract (WCE) (ySC7) on 1039-base-pair (bp) ARS1-containing DNAs. Bead-associated proteins were analyzed by immunoblotting. Sequences of wild-type and mutant A and B2 elements of ARS1 are shown. (B) Orc6 is required for stable association of Cdt1 in the presence of ATPγS. Assembly reactions were performed with ATP or ATPγS on 1039-bp B2 + M2 ARS1-containing DNA.
Previously, we found that ORCΔ6, but not complete ORC, is able to bind simultaneously to the ARS1 ACS and a second near match to the ACS at the ARS1 B2 element (Wilmes and Bell 2002). This finding raised the possibility that the Mcm2–7 loading defect observed with ORCΔ6 was caused by interference due to this additional binding event. To address this possibility, we repeated the same assays described above using an ARS1 B2 mutant (B2 + M2) that exhibits wild-type origin function but does not bind a second ORCΔ6 at B2 (Wilmes and Bell 2002). We observed the same ORCΔ6-dependent defect in Mcm2–7 loading using the B2 + M2 template (Fig. 4A, cf. lanes 4–6 and 7–9), indicating that ORCΔ6 binding to B2 was not responsible for the defects in pre-RC formation observed in the absence of Orc6.
ORCΔ6 is defective in recruiting Cdt1
What step in pre-RC assembly requires Orc6 function? The above analysis indicated that Orc6 was not required for recruitment of Cdc6 but was required for Mcm2–7 origin association. Cdt1 associates only transiently with origin DNA in the presence of ATP, and therefore is not detected in standard pre-RC assembly assays (Randell et al. 2006). To determine whether the Orc6 protein was required for Cdt1 association, we took advantage of the finding that inhibiting Cdc6 ATP hydrolysis with ATPγS stabilizes Cdt1 origin association while reducing Mcm2–7 loading (Fig. 4B, cf. lanes 1 and 2; Gillespie et al. 2001; Randell et al. 2006). In the presence of ATPγS, ORCΔ6 did not stabilize Cdt1 (Fig. 4B, cf. lanes 2 and 4). Addition of recombinant Orc6 to ORCΔ6 in the presence of ATPγS restored Cdt1 stabilization to the level of wild type ORC (Fig. 4B, cf. lanes 2 and 6). Thus, Orc6 is required to recruit Cdt1 to the origin.
Orc6 directly binds Orc3, Orc5, and Cdt1
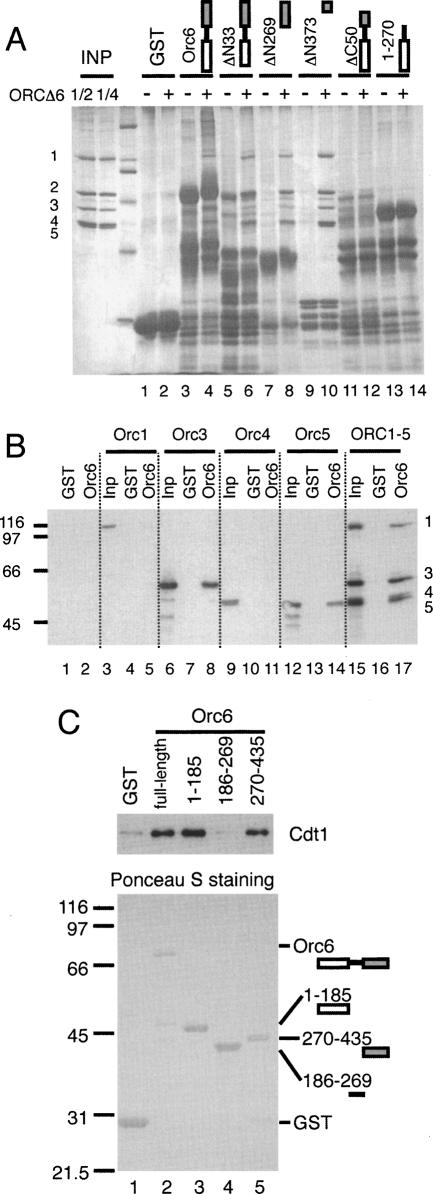
To investigate further the interactions between Orc6 and other pre-RC components, we expressed full-length and truncated forms of Orc6 in E. coli as GST fusion proteins and immobilized each on glutathione beads. To map the region of Orc6 that binds ORC, we added purified ORCΔ6 to each of the different forms of Orc6-beads and the total protein bound was analyzed by SDS-PAGE followed by silver staining. Both of the Orc6 C-terminal deletions, including one removing only 50 amino acids (ΔC50), lost the ability to bind ORCΔ6 (Fig. 5A, lanes 11–14). In contrast, N-terminal truncations removing all but the last 62 amino acids of Orc6 (ΔN373) retained the ability to bind ORCΔ6 (Fig. 5A, lanes 3–10). Thus, the C-terminal 62 residues of Orc6 (Orc6-CTD) are necessary and sufficient for Orc6 to bind ORCΔ6. Similar assays using individual ORC subunits show that Orc6 interacts with the Orc3 and Orc5 subunits but not Orc1 or Orc4 (Fig. 5B; Supplementary Fig. 3).
Figure 5.
Mapping of Orc6 protein-binding domains. (A) The C-terminal 62-amino-acid residues of Orc6 are sufficient for Orc1–5 binding. GST-Orc6 fusion proteins were bound to glutathione beads and washed before adding purified ORCΔ6. Total protein was assayed by silver staining after separation on SDS-PAGE. (B) Orc6 binds directly to Orc3 and Orc5. Individually expressed ORC subunits were incubated with bead-bound GST-Orc6 and were assayed by anti-Orc1, anti-Orc3, anti-Orc4, and anti-Orc5 Western analysis using antibodies SB13, SB3, SB6, and SB5, respectively. (C) Both the N- and C-terminal domains of Orc6 interact with Cdt1. GST-Orc6 fusion proteins were bound to glutathione beads and washed. (Top panel) Purified Cdt1 protein was incubated with GST-Orc6 and bound Cdt1 was assayed by immunoblotting. Ponceau S staining of glutathione bead-bound proteins is shown in the bottom panel.
To determine whether Orc6 binds Cdt1 directly, we tested the association of the GST-Orc6 fusions with purified Cdt1. Consistent with previous two-hybrid data (Semple et al. 2006), full-length Orc6 bound Cdt1 directly (Fig. 5C). Analysis of GST-Orc6 truncation constructs identified at least two regions that bind Cdt1 independently. Both the N-terminal 185 amino acids and the C-terminal 165 amino acids of Orc6 interacted strongly with Cdt1. In contrast, the middle region of Orc6 (residues 186–269) did not bind Cdt1. We also tested the ability of N-terminal deletion mutants (ΔN185, ΔN269) to function in the pre-RC assembly assay. Even though both mutants retained the ability to bind ORCΔ6, neither restored the ability to stabilize Cdt1 at the origin in the presence of ATPγS (data not shown).
Tethering Cdt1 to ORCΔ6 allows Mcm2–7 association in the absence of full-length Orc6
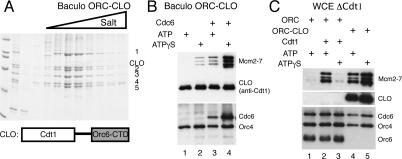
We hypothesized that Orc6 functions to recruit Cdt1 to origins through direct interactions with ORCΔ6 and Cdt1. To test this hypothesis, we tethered Cdt1 to ORCΔ6 using the 62-amino-acid, C-terminal domain of Orc6 (Orc6-CTD). For this purpose, we fused full-length Cdt1 to a flexible linker followed by the Orc6-CTD (Cdt1-linker-Orc6-CTD, hereafter called CLO). When coexpressed with ORCΔ6, CLO associated stoichiometrically with ORCΔ6 and these proteins copurified as a single complex (ORC–CLO) through multiple steps of chromatography (Fig. 6A).
Figure 6.
Tethering Cdt1 to ORCΔ6 can recruit Mcm2–7 in vitro. (A) ORC–CLO forms a single complex. CLO was coexpressed with Orc1–5 using baculovirus infection of insect cells and purified by ion exchange chromatography. A Coomassie-stained, SDS–polyacrylamide gel showing the coelution of Orc1–5 and the CLO protein from the second column (Mono-Q, GE Healthcare) is shown. CLO associates stoichiometrically with Orc1–5. (B) ORC–CLO binds to B2 + M2 ARS1 DNA and loads Mcm2–7 efficiently in the presence of Cdc6. CLO was detected using anti-Cdt1 antiserum HM5352 (Randell et al. 2006). Note: In a typical pre-RC assembly reaction, ORC is ∼10-fold more abundant than Cdt1, even when the latter is stabilized by ATPγS. This was the reason for the strong CLO signals in B and C, and in Figure 7. (C) Mcm2–7 loading by ORC–CLO does not depend on endogenous Cdt1. Pre-RC assembly assays were performed with ORC- and Cdt1-depleted WCE.
The CLO fusion protein restored the ability of ORCΔ6 to recruit Mcm2–7 proteins to origin DNA (Fig. 6B, cf. lanes 3 and 4). Cdt1 has affinity for MCM proteins (Tanaka and Diffley 2002b); however, this was not the basis of Mcm2–7 recruitment by ORC–CLO because in the presence of ATP ORC–CLO was unable to recruit Mcm2–7 in the absence of Cdc6 (Fig. 6B, cf. lanes 1 and 3). Cdc6-dependence is a hallmark of pre-RC assembly both in vitro and in vivo. Consistent with the hypothesis that the tethered form of Cdt1 (CLO) recruits Mcm2–7, extracts depleted of endogenous Cdt1 still showed robust Mcm2–7 recruitment in the presence of ORC–CLO and Cdc6 (Fig. 6C). In contrast, Mcm2–7 recruitment by wild-type ORC required addition of purified Cdt1 to the same depleted extracts (Fig. 6C, cf. lanes 1, 2, and 4).
ATP hydrolysis is critical to the control of pre-RC assembly, so we asked how inhibiting ATP hydrolysis with ATPγS affected Mcm2–7 recruitment by ORC–CLO. As observed previously, in the presence of wild-type ORC addition of ATPγS reduces Mcm2–7 association (see Fig. 4B). In contrast, addition of ATPγS to ORC–CLO reactions led to much higher levels of Mcm2–7 association (Fig. 6B). Because ATPγS inhibits repeated rounds of Mcm2–7 loading (Randell et al. 2006), these results suggest that recruitment of Cdt1 to ORC is limiting for the initial recruitment of Mcm2–7 to the origin. Intriguingly, in the presence of ATPγS but not ATP, a lesser, but clearly detectable, level of Mcm2–7 was recruited by ORC–CLO even in the absence of Cdc6 (Fig. 6B, cf. lanes 1 and 2). This finding indicates that Orc1–5 or the CLO protein interacts with the Mcm2–7 proteins directly when ATP hydrolysis is inhibited.
The properties of the ORC–CLO complex during pre-RC formation require the CLO protein to be incorporated into ORC. We tested the ability of the CLO protein to function in pre-RC assembly assays lacking Cdt1 protein (Supplementary Fig. 4). For these studies, we expressed CLO in E. coli separately from ORC. When CLO was added to Cdt1-depleted extracts containing the full ORC complex we observed no difference in Mcm2–7 association compared with addition of Cdt1. In both cases we saw robust Mcm2–7 association in the presence of ATP and reduced association in the presence of ATPγS. Thus, appending Orc6-CTD to Cdt1 did not eliminate its ability to perform the normal Cdt1 function in a pre-RC assay. In contrast, when we added CLO protein to an extract containing ORCΔ6, the pattern of Mcm2–7 association changed to mimic pre-RC assembly in the presence of ORC–CLO: Mcm2–7 association was reduced in the presence of ATP and elevated in the presence of ATPγS. Importantly, consistent with there being only a single binding site for Orc6-CTD on ORC, when we tested the ability of CLO to interact with ORC and ORCΔ6, we found that CLO can only bind to ORCΔ6 (Supplementary Fig. 5). Together, these experiments demonstrate that the distinct activity of ORC–CLO is due to the tethering of Cdt1 to ORC rather than a change in Cdt1 function when Orc6-CTD is attached to the C terminus of Cdt1.
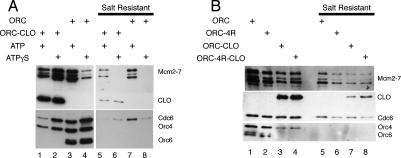
Mcm2–7 initially associates with origins via protein–protein interactions, followed by its loading onto origin DNA after Cdc6 ATP hydrolysis (Randell et al. 2006). To address whether ORC–CLO can load Mcm2–7 onto origin DNA, we used salt extraction to remove Mcm2–7 complexes that are associated with origin DNA via interactions with ORC and other pre-RC assembly factors (Bowers et al. 2004). After salt extraction, only “loaded” Mcm2–7 complexes, which are bound to the origin DNA directly rather than through ORC, are retained. Whether ORC or ORC–CLO was used in the pre-RC assembly assays, we found that a similar percentage of Mcm2–7 complexes were resistant to salt (Fig. 7A). In both cases, salt-resistant complexes are not formed in the presence of nonhydrolyzable ATPγS. Thus, although much higher levels of Mcm2–7 proteins can associate with ORC–CLO in the presence of ATPγS, these proteins are not loaded onto the origin (Fig. 7A, cf. lanes 2 and 6). Together, these findings show that ORC–CLO loads Mcm2–7 onto origin DNA in an ATP-hydrolysis-dependent manner.
Figure 7.
ORC–CLO can only perform one round of Mcm2–7 loading. (A) Mcm2–7 complexes loaded by ORC–CLO are resistant to salt extraction (cf. lane 5). Pre-RCs were formed with ORC or ORC–CLO in ORC-depleted extract as for Figure 6B, and were incubated in buffer H/500mM NaCl for salt extraction. (B) ORC–CLO cannot load Mcm2–7 complexes reiteratively. Comparison between ORC–CLO and its Orc4-R267A-containing derivative ORC-4R–CLO showed they both load same amount of Mcm2–7 (cf. lanes 7,8).
ORC-CLO cannot repeatedly load Mcm2–7 complexes
The ability of ORC–CLO to load Mcm2–7 onto origin DNA suggested that the CLO fusion gene might complement an ORC6 deletion. We tested this possibility and found that CLO did not complement strains lacking ORC6 at either low or high expression levels (data not shown). The identification of an ORC mutant that loaded Mcm2–7 at reduced levels and was unable to complement ORC function in vivo was reminiscent of our analysis of an allele of ORC4 (orc4-R267A) that inhibited ORC ATP hydrolysis (Bowers et al. 2004). An ORC complex including this mutant (ORC-4R) could catalyze one round of Mcm2–7 loading at an origin in vitro but was defective for reiterative Mcm2–7 loading.
To address whether ORC–CLO is capable of multiple rounds of Mcm2–7 loading, we replaced Orc4 with Orc4-R267A (ORC–CLO-4R) to inhibit ORC–CLO ATP hydrolysis and therefore allow only a single round of Mcm2–7 loading. If ORC-CLO ordinarily performs multiple rounds of Mcm2–7 loading, then the lack of ATP hydrolysis by ORC–CLO-4R should reduce the level of Mcm2–7 loading relative to ORC–CLO. Consistent with previous studies, ATPase assays with ORC–CLO and ORC–CLO-4R showed substantially reduced ATP hydrolysis in the presence of Orc4-R267A (Supplementary Table 2). Importantly, ORC–CLO and ORC hydrolyze ATP at the same rate, indicating that tethering Cdt1 to ORC does not affect ORC ATP hydrolysis.
We compared the levels of Mcm2–7 association (before salt extraction) and loading (after salt extraction) mediated by ORC, ORC–CLO, and the “4R” derivatives of both these complexes (Fig. 7B). As previously observed, ORC-4R reduced Mcm2–7 origin association (Fig. 7B, cf. lanes 1 and 2) and loading (Fig. 7B, cf. lanes 5 and 6) relative to wild-type ORC. In contrast, incorporating Orc4-R267A into ORC–CLO resulted in no change in the extent of Mcm2–7 association (Fig. 7B, cf. lanes 3 and 4) or loading (Fig. 7B, cf. lanes 7 and 8). Thus, tethering Cdt1 to ORC inhibited pre-RC formation prior to the first step that requires ORC ATP hydrolysis and before multiple Mcm2–7 complexes can be loaded. Together, these data support a model in which Cdt1 is required for the assembly of the first Mcm2–7 onto origin DNA, but Cdt1 release is required for additional rounds of Mcm2–7 loading to occur.
Discussion
Orc6 is the smallest ORC subunit and the only one that is unrelated to AAA+ ATPases. We used a combination of in vivo and in vitro analysis to dissect the function of this protein during the cell cycle. Using genome-wide assays for ORC DNA binding and pre-RC formation, we show that Orc6 is not required for ORC DNA binding in vivo, but is essential for assembly and maintenance of pre-RCs. In vitro studies of Orc6 reveal that this subunit recruits Cdt1 to the origin and that this function can be bypassed in vitro by tethering full-length Cdt1 to ORC lacking the majority of Orc6. Further studies of ORCΔ6 tethered to Cdt1 support a model in which Cdt1 binding and release is critical for the essential process of loading multiple Mcm2–7 complexes at the origin.
Elimination of Orc6 causes a global loss of pre-RC formation
Previous analysis of the effect of transcriptionally inhibiting Orc6 expression in G1 cells showed that there was a partial loss of Mcm2 association with chromatin when Orc6 levels were reduced (Semple et al. 2006). This partial effect could have been due to an Orc6 requirement for the maintenance of pre-RCs at only a subset of origins or incomplete depletion of Orc6. Our data indicate that Orc6 function is required for pre-RC formation and maintenance at all origins, and indicate that the partial effects of the previous analysis were likely due to residual Orc6 activity. In support of this conclusion, strains with transcriptionally repressed ORC6 continued to divide (Semple et al. 2006), whereas elimination of Orc6-td resulted in a rapid first cycle arrest of cell division (Supplementary Table 2).
Our in vivo studies do not definitively determine whether Orc6 functions during pre-RC formation or maintenance; however, our in vitro studies clearly demonstrate that Orc6 functions during pre-RC formation. Elimination of Orc6 inhibits the initial association of both Cdt1 and Mcm2–7, but not Cdc6, with origin DNA. Direct protein–protein interaction studies indicate that Orc6 contains at least two domains capable of binding Cdt1 and both of these domains are required for Orc6 function in vivo (data not shown). It is possible that these domains interact with different parts of Cdt1 since there is no strong homology between these regions of Orc6. Alternatively, Orc6 could recruit multiple Cdt1 molecules, which could, in turn, recruit multiple copies of the Mcm2–7 complex. Such a mechanism could allow the coordinated loading of pairs of Mcm2–7 complexes. In addition, phosphorylation of the Orc6 N-terminal domain is known to inhibit pre-RC formation (Nguyen et al. 2001), raising the possibility that this phosphorylation inhibits Cdt1 binding. We confirmed that the primary role of Orc6 during pre-RC formation was to recruit Cdt1 by tethering Cdt1 to the Orc1–5 subunits. Importantly, the resulting ORC-CLO complex showed the ability to recruit and load Mcm2–7 complexes onto origin DNA.
Although our findings clearly demonstrate that Orc6 is required to recruit Cdt1 during pre-RC formation, other proteins also participate in this event. Consistent with this conclusion, even in the presence of the full Orc6 protein, we do not observe recruitment of Cdt1 to the origin in the absence of Cdc6 (data not shown). Indeed, our previous studies have shown that Cdc6 must both be present and bound to ATP to observe stable Cdt1 association with the origin (Randell et al. 2006). At this point it is not clear whether the role of Cdc6 bound to ATP is to change the conformation of the ORC–Cdc6–origin complex to reveal the Orc6–Cdt1-binding site or to provide additional critical contacts with Cdt1 that depend on Cdc6 binding to ATP.
Although recruiting Cdt1 to the origin is clearly an essential function of Orc6, we cannot eliminate the possibility that Orc6 plays other important functions both later in replication or in other cellular events. Studies in other organisms show a clear role for Orc6 in cytokinesis (Prasanth et al. 2002; Chesnokov et al. 2003); however, we see no delay in passage through the M–G1 transition in cells depleted for Orc6 (see Fig. 2C; data not shown). It is also possible that Orc6 functions during the formation of other origin-associated protein complexes (e.g., the preinitiation complex) after pre-RC formation. Unfortunately, such a function of Orc6 is difficult to assess in vivo because elimination of Orc6 in late G1 leads to the disassembly of pre-RCs, which are required to form subsequent complexes at the origin.
The ORC–CLO complex allowed the detection of interactions between ORC and the Mcm2–7 complex. When ORC–CLO is assayed in the absence of Cdc6 we observed clear recruitment of Mcm2–7 proteins in the presence of ATPγS but not ATP (Fig. 6B, lanes 1,2). Because wild-type ORC does not show any association with the Mcm2–7 complex under any condition (Randell et al. 2006), this observation is consistent with Cdt1 facilitating Mcm2–7 recruitment. However, tethering Cdt1 to the DNA (via ORC) is not sufficient to recruit Mcm2–7 because this interaction is only observed when ATP-γ-S is present. These findings have two important implications. First, the interaction must involve ORC since Cdt1 is not regulated by ATP binding. Second, since the interaction is only observed in the presence of ATPγS, which mimics the ATP-bound state of ORC, the Mcm2–7 association requires the ATP-bound form of ORC and is released by ATP hydrolysis (thus, this association is not observed in reactions containing ATP when hydrolysis can occur). This observation supports a model in which ATP hydrolysis by ORC is involved in the release of Mcm2–7 from ORC at the end of a cycle of Mcm2–7 loading (Randell et al. 2006).
Dynamic interactions between Cdt1 and ORC
Our analysis of the ORC–CLO complex suggests that binding and release of Cdt1 is important for repetitive loading of the Mcm2–7 complex at origins. Although this tethered Cdt1 complex can load the Mcm2–7 complex onto the origin, ORC–CLO cannot repeat this process, which is essential for full pre-RC formation. The requirement for Cdt1 release for repetitive Mcm2–7 loading is consistent with observations indicating that Cdt1 associates dynamically with the origin. For example, Xenopus and S. cerevisiae studies both indicate that Cdt1 is stabilized at the origin by preventing ATP hydrolysis (Gillespie et al. 2001; Randell et al. 2006). Similarly, the inability to detect Cdt1 association with individual origins by ChIP (data not shown) is consistent with a dynamic association with the origin.
What is the role for Cdt1 binding and release? Because Cdt1 interacts with and stabilizes Mcm2–7 at the origin (see above), it is possible that Cdt1 release is required to reduce the affinity of the initial association of the Mcm2–7 complex with the ORC–Cdc6 complex. Under these circumstances, tethering Cdt1 to ORC would prevent release of the first Mcm2–7 complex and therefore inhibit binding of a second Mcm2–7 complex. The continuous presence of Cdt1 could also prevent ORC from achieving a conformation necessary for a new round of loading. In this regard, it is interesting that, in the presence of ATP, the ORC–CLO complex consistently associates with less Cdc6 than wild-type ORC (see Figs. 6, 7). This reduced association could represent an inability of ORC–CLO to return to a conformation that can interact with Cdc6. We note that this defect is not observed in ATPγS, suggesting that ORC–CLO does not have a generally reduced affinity for Cdc6.
Relevance to replication in other organisms
Although Orc6 is the least conserved ORC subunit, our findings have substantial implications for replication in all eukaryotes. The sequence of Orc6 has changed substantially over evolutionary time, but its essential role in replication has remained (Chesnokov et al. 2001, 2003; Prasanth et al. 2002). It is possible that a role for Orc6 in recruiting Cdt1 to the origin has been lost in more complex organisms; however, it is noteworthy that the Cdt1 sequence is the second most diverse when comparing the sequence of pre-RC components between different eukaryotes. The diversity of Orc6 and Cdt1 sequences raises the possibility that these proteins have coevolved to maintain their interactions. A number of studies indicate that Orc6 has taken on additional roles in DNA binding and cytokinesis in metazoan organisms. Perhaps the process of developing these additional roles drove changes in Orc6 sequence that required compensatory changes in Cdt1. Finally, the dynamic association of Cdt1 with the pre-RC is conserved (see above) and given the highly conserved nature of other pre-RC proteins (Orc1–5, Cdc6, Mcm2–7) and the pre-RC assembly process, it is likely that the requirement for the binding and release of Cdt1 serves the same function in all organisms.
Reconciling in vivo and in vitro Orc6 functions
The observation that Orc6 is required for pre-RC formation and maintenance has interesting implications. In vitro experiments and analysis of in vivo isolated chromatin indicate that, once loaded, Mcm2–7 complexes do not need ORC or Cdc6 to be retained on the DNA (Donovan et al. 1997; Rowles et al. 1999; Edwards et al. 2002; Bowers et al. 2004). In contrast, analyses of mutations in ORC subunits and Cdc6 indicate that these proteins are necessary to maintain Mcm2–7 at the origin in vivo (Aparicio et al. 1997; Semple et al. 2006; this study). A hypothesis that would reconcile these data would be to invoke the presence of an activity that removes Mcm2–7 from origins during G1. Under normal conditions, the pre-RC assembly machinery would act to overcome this activity. However, if assembly was inhibited, the unloading activity would dominate. If correct, this model would predict that any protein required for pre-RC formation would also be required for its maintenance in G1. This is true for ORC and Cdc6, and it will be interesting to test if the same is true for Cdt1. It is also possible that one of the proteins required for loading Mcm2–7 complexes is also required for their release. Given the sequence and functional similarity of ORC and Cdc6 to sliding clamp loading proteins (Perkins and Diffley 1998; Randell et al. 2006), it is interesting to note that a subset of the proteins required to load sliding clamps are sufficient to unload clamps (Leu et al. 2000; Yao et al. 2006). Perhaps a subset of the pre-RC loading machinery is capable of unloading Mcm2–7 complexes from the DNA.
Materials and methods
Yeast strain construction
The BAR1 gene in yKL204 (Sanchez-Diaz et al. 2004) was disrupted with LEU2 by transforming the XbaI digestion of pZV77 (ySC167) to facilitate α-factor arrest. In galactose medium, this strain overexpresses the Ubr1 protein that recognizes the DHFR degradation signal. The Orc6 degron strain (ySC166) was generated by a one-step PCR method to introduce a heat-inducible degron cassette (Labib et al. 2000) at the N terminus of ORC6 in the parental strain ySC167. Briefly, when integrated, this construct fuses an ubiquitin-coupled temperature-sensitive form of the DHFR protein to the ORC6 ORF. The resulting ORF is expressed from the copper-inducible CUP1 promoter. Orc6 degradation was induced by growth at elevated temperature (to inactivate DHFR) in galactose (to induce Ubr1) and in the absence of Cu2+ (to repress the CUP1 promoter). See Supplementary Table 3 for a complete list of the strains used and their genotypes.
ChIP assays
ChIP assays were performed as described previously (Aparicio et al. 1997). Thirty-three milliliters of culture were collected for each ChIP sample. One-third of the resulting cell lysate was used for Mcm2–7 IP using an anti-Mcm2–7 polyclonal antibody (UM185), and one-third was used for ORC IP using an anti-ORC polyclonal antibody (1108).
Genome-wide location analyses
Half of the immunoprecipitated DNA and 5 μg of ChIP input DNA were differentially labeled with 2 nmol of either Cy3-dUTP or Cy5-dUTP (GE Healthcare) using 4 μg of random nonomer oligo (IDT) and 0.5 μL of 50,000 U/mL Klenowexo− (New England Biolabs) in a 40-μL reaction. IP and input samples were mixed after labeling, and unincorporated dyes were removed using a microcon filter (Millipore YM-30) by washing the samples three times with 0.4 mL of TE. The labeled DNAs were hybridized to custom-made DNA arrays from Agilent Technologies containing 44,290 probes spanning the yeast genome using standard protocol for cDNA hybridization and washing.
The resulting slides were scanned with an Agilent scanner. For each cohybridization, Cy3 and Cy5 levels were calculated using Agilent Feature Extractor CGH software. Log2 ratios for each experiment and scale normalizations across each set of duplicated experiments were calculated using the sma package (Yang et al. 2002) in R, a computer language and environment for statistical computing (version 2.1.0, http://www.r-project.org). Each averaged set of experiments included a biological duplicate and a dye swap in labeling. Averaged and raw data sets as well as a full description of the microarrays are available online in the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo, accession no. GSE9318).
Plasmid construction
Full-length and truncations of the ORC6 ORF were amplified with oligos containing EcoRI and XhoI sites and were cloned into the same sites in GST fusion vector pGEX-6p-3 (GE Healthcare). Ligation reactions were transformed into Top10F′ competent cells (Invitrogen) and were selected on LB + Amp at room temperature.
Cdt1-LNK-ORC6-CTD was cloned by first introducing ORC6-CTD to the 3′ end of the Cdt1 ORF followed by insertion of a 24-amino-acid flexible linker [R(SGGGG)4SGS] into the BglII site between Cdt1 and ORC6-CTD. This construct was cloned into pFastBacDual (BD Biosciences) for expression in insect cells. The Bacmid of Cdt1-LNK-ORC6-CTD was generated from DH10Bac cells transformed with pFastBacDual-Cdt1-LNK-ORC6-CTD, which underwent homologous recombination with the baculovirus genome.
Protein expression and purification
Wild-type and mutant ORC and Orc1–5 were expressed in insect cells and purified as described (Klemm et al. 1997). ORC–CLO was expressed in insect cells (HighFive, Invitrogen) by coinfection with Orc1–5 and Cdt1-LNK-ORC6-CTD viruses. Cells were collected 40 h post-infection, and a 10%–50% ammonium sulfate cut of the nuclear extract was subjected to purification on a 5-mL HiTrap SP column and a 1-mL MonoQ column (GE Healthcare). Single ORC subunits were expressed in insect cells upon baculovirus infection and were partially purified as supernatants of a 10% ammonium sulfate cut of the nuclei fraction. GST-Orc6 was expressed in Rossetta2(DE3)pLysS cells (Novagen) by transforming with pGEX-6p-3-ORC6 and was induced at room temperature with 1 mM IPTG at OD600 ∼0.6 for 4 h. The fusion protein was bound to glutathione Sepharose beads (GE Healthcare) in 50 mM HEPES (pH 7.6), 5 mM Mg-acetate, 1 mM EDTA, 1 mM EGTA, 20% glycerol, 0.05% NP-40, and 400 mM KCl, and was washed with 50 mM HEPES (pH 7.6), 5 mM Mg-acetate, 1 mM EDTA, 1 mM EGTA, 10% glycerol, 0.05% NP-40, and 150 mM KCl. Orc6 was cleaved off glutathione Sepharose beads with PreScission protease (GE Healthcare). Cdc6 and Cdt1 were expressed and purified as described previously (Randell et al. 2006).
GST pull-down assays
Lysates of GST-Orc6 and GST-Orc6 fragments were incubated with 2.5 μL of glutathione Sepharose beads as described above, and ∼1 μg of fusion protein was bound. The Orc-bound glutathione Sepharose beads were washed with 0.25 mL of H150 three times and were resuspended in 50 μL of H150 before incubation with 0.75 μg of purified Cdt1, 4 μg of purified ORCΔ6, or individually expressed ORC subunits from baculovirus-infected insect cells for 4 h at 4°C. Beads were washed with 0.25 mL of H150 three times and eluted in 0.2 mL of H150 containing 10 mM glutathione. One-fourth of the total proteins was assayed on a 10% SDS-PAGE.
Preparation of whole-cell extracts (WCEs) and ORC-depleted extracts
Yeast WCEs from strain ySC7 were prepared as described (Bowers et al. 2004, Randell et al. 2006). Cells were grown in YPD to an OD600 of 0.6, arrested in α factor for 4 h, and collected by centrifugation. To deplete ORC from extracts, 0.4 mL of ySC7 WCE was passed through two sequential Vivapure Mini-H S-Sepharose spin columns as described (Bowers et al. 2004).
Preparation of ARS1-DNA-coupled beads
ARS1-containing DNA substrates were generated by PCR using Advantage 2 Polymerase (BD Biosciences) and the primers RSP23 and ARS1-3Bio with plasmids pARS1/WT (WT) or pARS1/A−B2− (A−B2−) or the primers GWpARS1rev and ARS1-3Bio with plasmid pARS1/B2 + M2/ADE2 (B2 + M2). The DNA substrates were either purified with a Vivaspin 4-mL concentrator or on a 1% agarose gel. The ARS1-containing DNA was coupled to streptavidin-coated magnetic beads (Dynal) by incubation overnight at 1 pmol DNA/0.1 mg beads, and unbound DNA was removed by two washes with buffer H/300 mM potassium glutamate.
Pre-RC assembly assays
Pre-RC assembly assays used 0.5 pmol 1039-base-pair ARS1 DNA coupled to magnetic beads with 2 pmol recombinant ORC or ORCΔ6 or 4 pmol ORC-CLO, 8 pmol recombinant Cdc6, and 10 μL of ORC-depleted ySC7 WCE in 20-μL reactions and were incubated for 20 min at room temperature as described previously (Bowers et al. 2004; Randell et al. 2006). Salt extraction experiments were carried out by washing assembled pre-RC twice with buffer H/500 mM NaCl for 1 min. Cdt1 immunodepletion was performed as described previously (Randell et al. 2006). Pre-RC components were analyzed by immunoblotting using antiserum UM185 (Mcm2–7), HM5352 (Cdt1), and monoclonal antibodies SB6 and SB12 (Orc4), SB49 (Orc6), and 12CA5 (HA tag).
Acknowledgments
We thank Karim Labib for providing yeast strain yKL204 and degron construct plasmid pKL187; Ray Deshaies for providing pZV77; Hannah Blitzblau and Andy Fan for help with DNA microarrays; and Tania Baker, Hannah Blitzblau, Alan Grossman, Ryan Heller, Michael Laub, David MacAlpine, John C.W. Randell, Jue Wang, and Vasiliki Tsakraklides, as well as the reviewers, for critical reading and helpful comments on this manuscript. The work described here was supported by a grant from the National Institute of Health to S.P.B. (GM52339). S.P.B. and S.C. are employees of the Howard Hughes Medical Institute. S.C. was supported by a fellowship from the Anna Fuller Fund.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1596807
References
- Aparicio O.M., Weinstein D.M., Bell S.P., Weinstein D.M., Bell S.P., Bell S.P. Components and dynamics of DNA replication complexes in S. cerevisiae: Redistribution of MCM proteins and Cdc45p during S phase. Cell. 1997;91:59–69. doi: 10.1016/s0092-8674(01)80009-x. [DOI] [PubMed] [Google Scholar]
- Balasov M., Huijbregts R.P., Chesnokov I., Huijbregts R.P., Chesnokov I., Chesnokov I. Role of the Orc6 protein in origin recognition complex-dependent DNA binding and replication in Drosophila melanogaster. Mol. Cell. Biol. 2007;27:3143–3153. doi: 10.1128/MCB.02382-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers J.L., Randell J.C.W., Chen S., Bell S.P., Randell J.C.W., Chen S., Bell S.P., Chen S., Bell S.P., Bell S.P. ATP hydrolysis by ORC catalyzes reiterative Mcm2–7 assembly at a defined origin of replication. Mol. Cell. 2004;16:967–978. doi: 10.1016/j.molcel.2004.11.038. [DOI] [PubMed] [Google Scholar]
- Chesnokov I., Remus D., Botchan M., Remus D., Botchan M., Botchan M. Functional analysis of mutant and wild-type Drosophila origin recognition complex. Proc. Natl. Acad. Sci. 2001;98:11997–12002. doi: 10.1073/pnas.211342798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesnokov I.N., Chesnokova O.N., Botchan M., Chesnokova O.N., Botchan M., Botchan M. A cytokinetic function of Drosophila ORC6 protein resides in a domain distinct from its replication activity. Proc. Natl. Acad. Sci. 2003;100:9150–9155. doi: 10.1073/pnas.1633580100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da-Silva L.F., Duncker B.P., Duncker B.P. ORC function in late G1: Maintaining the license for DNA replication. Cell Cycle. 2007;6:128–130. doi: 10.4161/cc.6.2.3743. [DOI] [PubMed] [Google Scholar]
- Donovan S., Harwood J., Drury L.S., Diffley J.F.X., Harwood J., Drury L.S., Diffley J.F.X., Drury L.S., Diffley J.F.X., Diffley J.F.X. Cdc6p-dependent loading of Mcm proteins onto pre-replicative chromatin in budding yeast. Proc. Natl. Acad. Sci. 1997;94:5611–5616. doi: 10.1073/pnas.94.11.5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards M.C., Tutter A.V., Cvetic C., Gilbert C.H., Prokhorova T.A., Walter J.C., Tutter A.V., Cvetic C., Gilbert C.H., Prokhorova T.A., Walter J.C., Cvetic C., Gilbert C.H., Prokhorova T.A., Walter J.C., Gilbert C.H., Prokhorova T.A., Walter J.C., Prokhorova T.A., Walter J.C., Walter J.C. MCM2–7 complexes bind chromatin in a distributed pattern surrounding the origin recognition complex in Xenopus egg extracts. J. Biol. Chem. 2002;277:33049–33057. doi: 10.1074/jbc.M204438200. [DOI] [PubMed] [Google Scholar]
- Ekholm-Reed S., Mendez J., Tedesco D., Zetterberg A., Stillman B., Reed S.I., Mendez J., Tedesco D., Zetterberg A., Stillman B., Reed S.I., Tedesco D., Zetterberg A., Stillman B., Reed S.I., Zetterberg A., Stillman B., Reed S.I., Stillman B., Reed S.I., Reed S.I. Deregulation of cyclin E in human cells interferes with prereplication complex assembly. J. Cell Biol. 2004;165:789–800. doi: 10.1083/jcb.200404092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erzberger J.P., Berger J.M., Berger J.M. Evolutionary relationships and structural mechanisms of AAA+ proteins. Annu. Rev. Biophys. Biomol. Struct. 2006;35:93–114. doi: 10.1146/annurev.biophys.35.040405.101933. [DOI] [PubMed] [Google Scholar]
- Gillespie P., Li A., Blow J.J., Li A., Blow J.J., Blow J.J. Reconstitution of licensed replication origins on Xenopus sperm nuclei using purified proteins. BMC Biochem. 2001;2:15. doi: 10.1186/1471-2091-2-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemm R.D., Austin R.J., Bell S.P., Austin R.J., Bell S.P., Bell S.P. Coordinate binding of ATP and origin DNA regulates the ATPase activity of the origin recognition complex. Cell. 1997;88:493–502. doi: 10.1016/s0092-8674(00)81889-9. [DOI] [PubMed] [Google Scholar]
- Labib K., Tercero J.A., Diffley J.F., Tercero J.A., Diffley J.F., Diffley J.F. Uninterrupted MCM2–7 function required for DNA replication fork progression. Science. 2000;288:1643–1647. doi: 10.1126/science.288.5471.1643. [DOI] [PubMed] [Google Scholar]
- Lee D., Bell S., Bell S. Architecture of the yeast origin recognition complex bound to origins of DNA replication. Mol. Cell. Biol. 1997;17:7159–7168. doi: 10.1128/mcb.17.12.7159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengronne A., Schwob E., Schwob E. The yeast CDK inhibitor Sic1 prevents genomic instability by promoting replication origin licensing in late G(1) Mol. Cell. 2002;9:1067–1078. doi: 10.1016/s1097-2765(02)00513-0. [DOI] [PubMed] [Google Scholar]
- Leu F.P., Hingorani M.M., Turner J., O’Donnell M., Hingorani M.M., Turner J., O’Donnell M., Turner J., O’Donnell M., O’Donnell M. The δ subunit of DNA polymerase III holoenzyme serves as a sliding clamp unloader in Escherichia coli. J. Biol. Chem. 2000;275:34609–34618. doi: 10.1074/jbc.M005495200. [DOI] [PubMed] [Google Scholar]
- Mizushima T., Takahashi N., Stillman B., Takahashi N., Stillman B., Stillman B. Cdc6p modulates the structure and DNA binding activity of the origin recognition complex in vitro. Genes & Dev. 2000;14:1631–1641. [PMC free article] [PubMed] [Google Scholar]
- Nguyen V.Q., Co C., Li J.J., Co C., Li J.J., Li J.J. Cyclin-dependent kinases prevent DNA re-replication through multiple mechanisms. Nature. 2001;411:1068–1073. doi: 10.1038/35082600. [DOI] [PubMed] [Google Scholar]
- Perkins G., Diffley J.F.X., Diffley J.F.X. Nucleotide-dependent prereplicative complex assembly by Cdc6p, a homolog of eukaryotic and prokaryotic clamp-loaders. Mol. Cell. 1998;2:23–32. doi: 10.1016/s1097-2765(00)80110-0. [DOI] [PubMed] [Google Scholar]
- Prasanth S.G., Prasanth K.V., Stillman B., Prasanth K.V., Stillman B., Stillman B. Orc6 involved in DNA replication, chromosome segregation, and cytokinesis. Science. 2002;297:1026–1031. doi: 10.1126/science.1072802. [DOI] [PubMed] [Google Scholar]
- Randell J.C., Bowers J.L., Rodriguez H.K., Bell S.P., Bowers J.L., Rodriguez H.K., Bell S.P., Rodriguez H.K., Bell S.P., Bell S.P. Sequential ATP hydrolysis by Cdc6 and ORC directs loading of the Mcm2–7 helicase. Mol. Cell. 2006;21:29–39. doi: 10.1016/j.molcel.2005.11.023. [DOI] [PubMed] [Google Scholar]
- Rowles A., Tada S., Blow J., Tada S., Blow J., Blow J. Changes in association of the Xenopusorigin recognition complex with chromatin on licensing of replication origins. J. Cell Sci. 1999;112:2011–2018. doi: 10.1242/jcs.112.12.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Diaz A., Kanemaki M., Marchesi V., Labib K., Kanemaki M., Marchesi V., Labib K., Marchesi V., Labib K., Labib K. Rapid depletion of budding yeast proteins by fusion to a heat-inducible degron. Sci. STKE. 2004;2004:PL8. doi: 10.1126/stke.2232004p18. [DOI] [PubMed] [Google Scholar]
- Seki T., Diffley J.F.X., Diffley J.F.X. Stepwise assembly of initiation proteins at budding yeast replication origins in vitro. Proc. Natl. Acad. Sci. 2000;97:14115–14120. doi: 10.1073/pnas.97.26.14115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semple J.W., Da-Silva L.F., Jervis E.J., Ah-Kee J., Al-Attar H., Kummer L., Heikkila J.J., Pasero P., Duncker B.P., Da-Silva L.F., Jervis E.J., Ah-Kee J., Al-Attar H., Kummer L., Heikkila J.J., Pasero P., Duncker B.P., Jervis E.J., Ah-Kee J., Al-Attar H., Kummer L., Heikkila J.J., Pasero P., Duncker B.P., Ah-Kee J., Al-Attar H., Kummer L., Heikkila J.J., Pasero P., Duncker B.P., Al-Attar H., Kummer L., Heikkila J.J., Pasero P., Duncker B.P., Kummer L., Heikkila J.J., Pasero P., Duncker B.P., Heikkila J.J., Pasero P., Duncker B.P., Pasero P., Duncker B.P., Duncker B.P. An essential role for Orc6 in DNA replication through maintenance of pre-replicative complexes. EMBO J. 2006;25:5150–5158. doi: 10.1038/sj.emboj.7601391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shima N., Alcaraz A., Liachko I., Buske T.R., Andrews C.A., Munroe R.J., Hartford S.A., Tye B.K., Schimenti J.C., Alcaraz A., Liachko I., Buske T.R., Andrews C.A., Munroe R.J., Hartford S.A., Tye B.K., Schimenti J.C., Liachko I., Buske T.R., Andrews C.A., Munroe R.J., Hartford S.A., Tye B.K., Schimenti J.C., Buske T.R., Andrews C.A., Munroe R.J., Hartford S.A., Tye B.K., Schimenti J.C., Andrews C.A., Munroe R.J., Hartford S.A., Tye B.K., Schimenti J.C., Munroe R.J., Hartford S.A., Tye B.K., Schimenti J.C., Hartford S.A., Tye B.K., Schimenti J.C., Tye B.K., Schimenti J.C., Schimenti J.C. A viable allele of Mcm4 causes chromosome instability and mammary adenocarcinomas in mice. Nat. Genet. 2007;39:93–98. doi: 10.1038/ng1936. [DOI] [PubMed] [Google Scholar]
- Speck C., Chen Z., Li H., Stillman B., Chen Z., Li H., Stillman B., Li H., Stillman B., Stillman B. ATPase-dependent cooperative binding of ORC and Cdc6 to origin DNA. Nat. Struct. Mol. Biol. 2005;12:965–971. doi: 10.1038/nsmb1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S., Diffley J.F., Diffley J.F. Deregulated G1-cyclin expression induces genomic instability by preventing efficient pre-RC formation. Genes & Dev. 2002a;16:2639–2649. doi: 10.1101/gad.1011002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S., Diffley J.F.X., Diffley J.F.X. Interdependent nuclear accumulation of budding yeast Cdt1 and Mcm2–7 during G1 phase. Nat. Cell Biol. 2002b;4:198–207. doi: 10.1038/ncb757. [DOI] [PubMed] [Google Scholar]
- Wilmes G.M., Bell S.P., Bell S.P. The B2 element of the Saccharomyces cerevisiae ARS1 origin of replication requires specific sequences to facilitate pre-RC formation. Proc. Natl. Acad. Sci. 2002;99:101–106. doi: 10.1073/pnas.012578499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y.H., Dudoit S., Luu P., Lin D.M., Peng V., Ngai J., Speed T.P., Dudoit S., Luu P., Lin D.M., Peng V., Ngai J., Speed T.P., Luu P., Lin D.M., Peng V., Ngai J., Speed T.P., Lin D.M., Peng V., Ngai J., Speed T.P., Peng V., Ngai J., Speed T.P., Ngai J., Speed T.P., Speed T.P. Normalization for cDNA microarray data: A robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 2002;30:e15. doi: 10.1093/nar/30.4.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao N.Y., Johnson A., Bowman G.D., Kuriyan J., O’Donnell M., Johnson A., Bowman G.D., Kuriyan J., O’Donnell M., Bowman G.D., Kuriyan J., O’Donnell M., Kuriyan J., O’Donnell M., O’Donnell M. Mechanism of proliferating cell nuclear antigen clamp opening by replication factor C. J. Biol. Chem. 2006;281:17528–17539. doi: 10.1074/jbc.M601273200. [DOI] [PubMed] [Google Scholar]