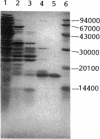
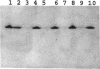
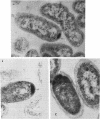
Abstract
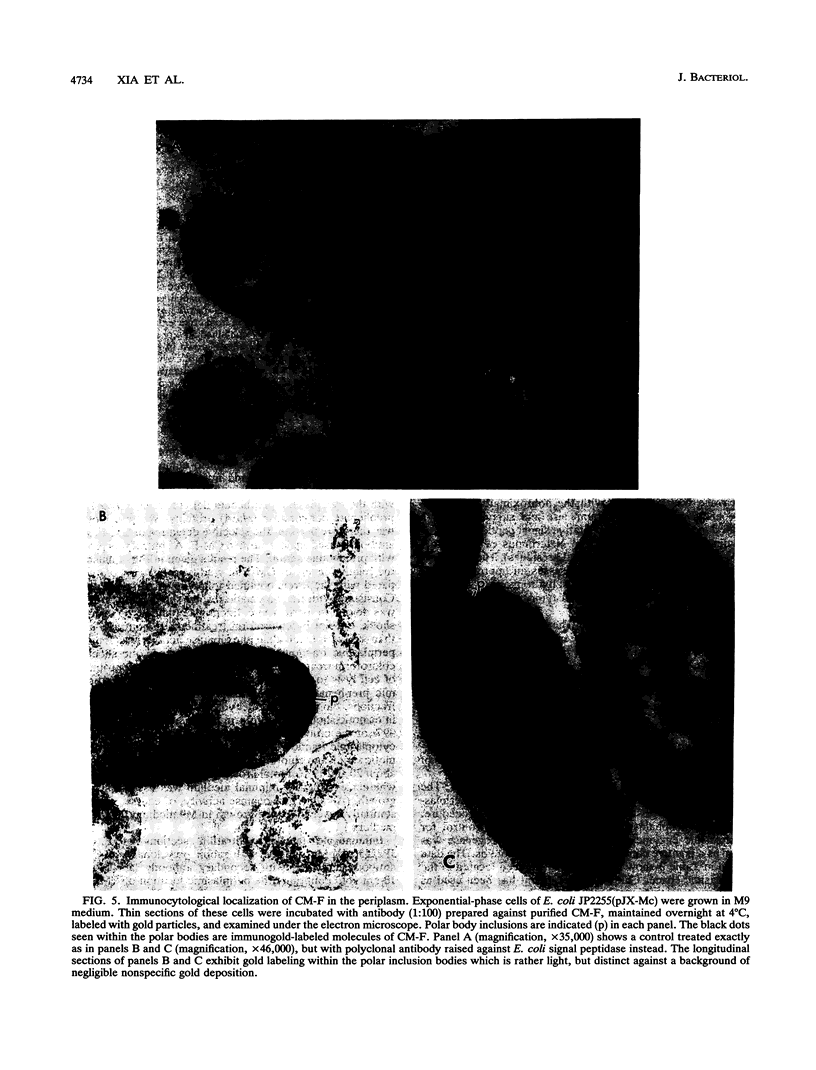
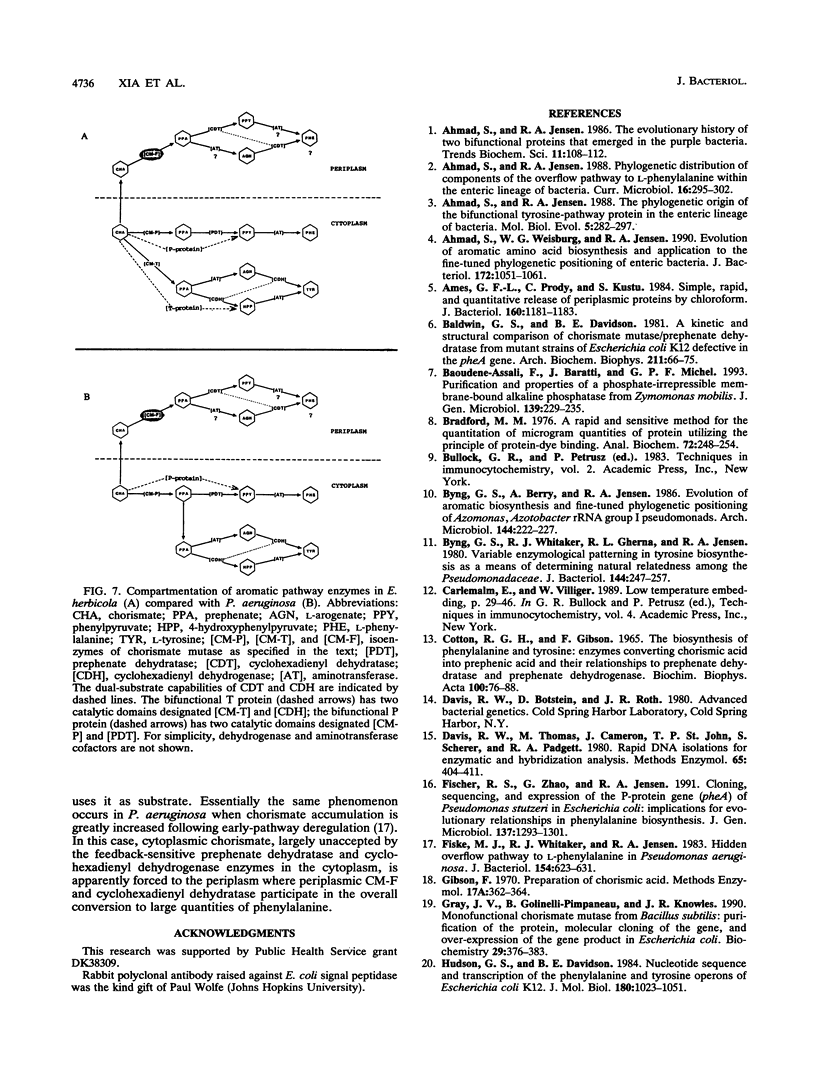
Enteric bacteria possess two species of chorismate mutase which exist as catalytic domains on the amino termini of the bifunctional PheA and TyrA proteins. In addition, some of these organisms possess a third chorismate mutase, CM-F, which exists as a small monofunctional protein. The CM-F gene (denoted aroQ) from Erwinia herbicola was cloned and sequenced for the first time. A strategy for selection by functional complementation in a chorismate mutase-free Escherichia coli background was devised by using a recombinant plasmid derivative of pUC18 carrying a Zymomonas mobilis tyrC insert which encodes cyclohexadienyl dehydrogenase. The aroQ gene is 543 bp in length, predicting a 181-residue protein product having a calculated molecular mass of 20,299 Da. The E. herbicola aroQ promoter is recognized by E. coli, and a putative sigma-70 promoter region was identified. N-terminal amino acid sequencing of the purified CM-F protein indicated cleavage of a 20-residue signal peptide. This was consistent with the monomeric molecular mass determined for the enzyme of about 18,000 Da. The native enzyme is a homodimer. The implied translocation of CM-F was confirmed by osmotic shock experiments which demonstrated a periplasmic location. Immunogold electron microscopy indicated a polar localization within the periplasm. Polyclonal antibody raised against E. herbicola CM-F did not cross-react with the CM-F protein from the closely related Serratia rubidaea, as well as from a number of other gram-negative bacteria. Furthermore, when the E. herbicola aroQ gene was used as a probe in Southern blot hybridizations with EcroRI digests of chromosomal DNA from S. rubidaea and other enteric organisms, no hybridization was detected at low stringency. Thus, the aroQ gene appears to be unusually divergent among closely related organisms. The deduced CM-F amino acid sequence did not exhibit compelling evidence for homology with the monofunctional chorismate mutase protein of Bacillus subtilis.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ahmad S., Jensen R. A. The phylogenetic origin of the bifunctional tyrosine-pathway protein in the enteric lineage of bacteria. Mol Biol Evol. 1988 May;5(3):282–297. doi: 10.1093/oxfordjournals.molbev.a040496. [DOI] [PubMed] [Google Scholar]
- Ahmad S., Weisburg W. G., Jensen R. A. Evolution of aromatic amino acid biosynthesis and application to the fine-tuned phylogenetic positioning of enteric bacteria. J Bacteriol. 1990 Feb;172(2):1051–1061. doi: 10.1128/jb.172.2.1051-1061.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ames G. F., Prody C., Kustu S. Simple, rapid, and quantitative release of periplasmic proteins by chloroform. J Bacteriol. 1984 Dec;160(3):1181–1183. doi: 10.1128/jb.160.3.1181-1183.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin G. S., Davidson B. E. A kinetic and structural comparison of chorismate mutase/prephenate dehydratase from mutant strains of Escherichia coli K 12 defective in the PheA gene. Arch Biochem Biophys. 1981 Oct 1;211(1):66–75. doi: 10.1016/0003-9861(81)90430-6. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Byng G. S., Whitaker R. J., Gherna R. L., Jensen R. A. Variable enzymological patterning in tyrosine biosynthesis as a means of determining natural relatedness among the Pseudomonadaceae. J Bacteriol. 1980 Oct;144(1):247–257. doi: 10.1128/jb.144.1.247-257.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- COTTON R. G., GIBSON F. THE BIOSYNTHESIS OF PHENYLALANINE AND TYROSINE; ENZYMES CONVERTING CHORISMIC ACID INTO PREPHENIC ACID AND THEIR RELATIONSHIPS TO PREPHENATE DEHYDRATASE AND PREPHENATE DEHYDROGENASE. Biochim Biophys Acta. 1965 Apr 12;100:76–88. doi: 10.1016/0304-4165(65)90429-0. [DOI] [PubMed] [Google Scholar]
- Davis R. W., Thomas M., Cameron J., St John T. P., Scherer S., Padgett R. A. Rapid DNA isolations for enzymatic and hybridization analysis. Methods Enzymol. 1980;65(1):404–411. doi: 10.1016/s0076-6879(80)65051-4. [DOI] [PubMed] [Google Scholar]
- Fischer R. S., Zhao G., Jensen R. A. Cloning, sequencing, and expression of the P-protein gene (pheA) of Pseudomonas stutzeri in Escherichia coli: implications for evolutionary relationships in phenylalanine biosynthesis. J Gen Microbiol. 1991 Jun;137(6):1293–1301. doi: 10.1099/00221287-137-6-1293. [DOI] [PubMed] [Google Scholar]
- Fiske M. J., Whitaker R. J., Jensen R. A. Hidden overflow pathway to L-phenylalanine in Pseudomonas aeruginosa. J Bacteriol. 1983 May;154(2):623–631. doi: 10.1128/jb.154.2.623-631.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray J. V., Golinelli-Pimpaneau B., Knowles J. R. Monofunctional chorismate mutase from Bacillus subtilis: purification of the protein, molecular cloning of the gene, and overexpression of the gene product in Escherichia coli. Biochemistry. 1990 Jan 16;29(2):376–383. doi: 10.1021/bi00454a011. [DOI] [PubMed] [Google Scholar]
- Hudson G. S., Davidson B. E. Nucleotide sequence and transcription of the phenylalanine and tyrosine operons of Escherichia coli K12. J Mol Biol. 1984 Dec 25;180(4):1023–1051. doi: 10.1016/0022-2836(84)90269-9. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Neu H. C., Heppel L. A. The release of enzymes from Escherichia coli by osmotic shock and during the formation of spheroplasts. J Biol Chem. 1965 Sep;240(9):3685–3692. [PubMed] [Google Scholar]
- Ozenberger B. A., Brickman T. J., McIntosh M. A. Nucleotide sequence of Escherichia coli isochorismate synthetase gene entC and evolutionary relationship of isochorismate synthetase and other chorismate-utilizing enzymes. J Bacteriol. 1989 Feb;171(2):775–783. doi: 10.1128/jb.171.2.775-783.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel N., Pierson D. L., Jensen R. A. Dual enzymatic routes to L-tyrosine and L-phenylalanine via pretyrosine in Pseudomonas aeruginosa. J Biol Chem. 1977 Aug 25;252(16):5839–5846. [PubMed] [Google Scholar]
- Prober J. M., Trainor G. L., Dam R. J., Hobbs F. W., Robertson C. W., Zagursky R. J., Cocuzza A. J., Jensen M. A., Baumeister K. A system for rapid DNA sequencing with fluorescent chain-terminating dideoxynucleotides. Science. 1987 Oct 16;238(4825):336–341. doi: 10.1126/science.2443975. [DOI] [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R. Bacterial evolution. Microbiol Rev. 1987 Jun;51(2):221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia T., Jensen R. A. Monofunctional chorismate mutase from Serratia rubidaea: a paradigm system for the three-isozyme gene family of enteric bacteria. Arch Biochem Biophys. 1992 Apr;294(1):147–153. doi: 10.1016/0003-9861(92)90149-q. [DOI] [PubMed] [Google Scholar]
- Xia T., Zhao G., Fischer R. S., Jensen R. A. A monofunctional prephenate dehydrogenase created by cleavage of the 5' 109 bp of the tyrA gene from Erwinia herbicola. J Gen Microbiol. 1992 Jul;138(7):1309–1316. doi: 10.1099/00221287-138-7-1309. [DOI] [PubMed] [Google Scholar]
- Xia T., Zhao G., Jensen R. A. The pheA/tyrA/aroF region from Erwinia herbicola: an emerging comparative basis for analysis of gene organization and regulation in enteric bacteria. J Mol Evol. 1993 Feb;36(2):107–120. doi: 10.1007/BF00166246. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Zhao G., Xia T., Aldrich H., Jensen R. A. Cyclohexadienyl dehydratase from Pseudomonas aeruginosa is a periplasmic protein. J Gen Microbiol. 1993 Apr;139(4):807–813. doi: 10.1099/00221287-139-4-807. [DOI] [PubMed] [Google Scholar]
- Zhao G., Xia T., Ingram L. O., Jensen R. A. An allosterically insensitive class of cyclohexadienyl dehydrogenase from Zymomonas mobilis. Eur J Biochem. 1993 Feb 15;212(1):157–165. doi: 10.1111/j.1432-1033.1993.tb17646.x. [DOI] [PubMed] [Google Scholar]