Abstract
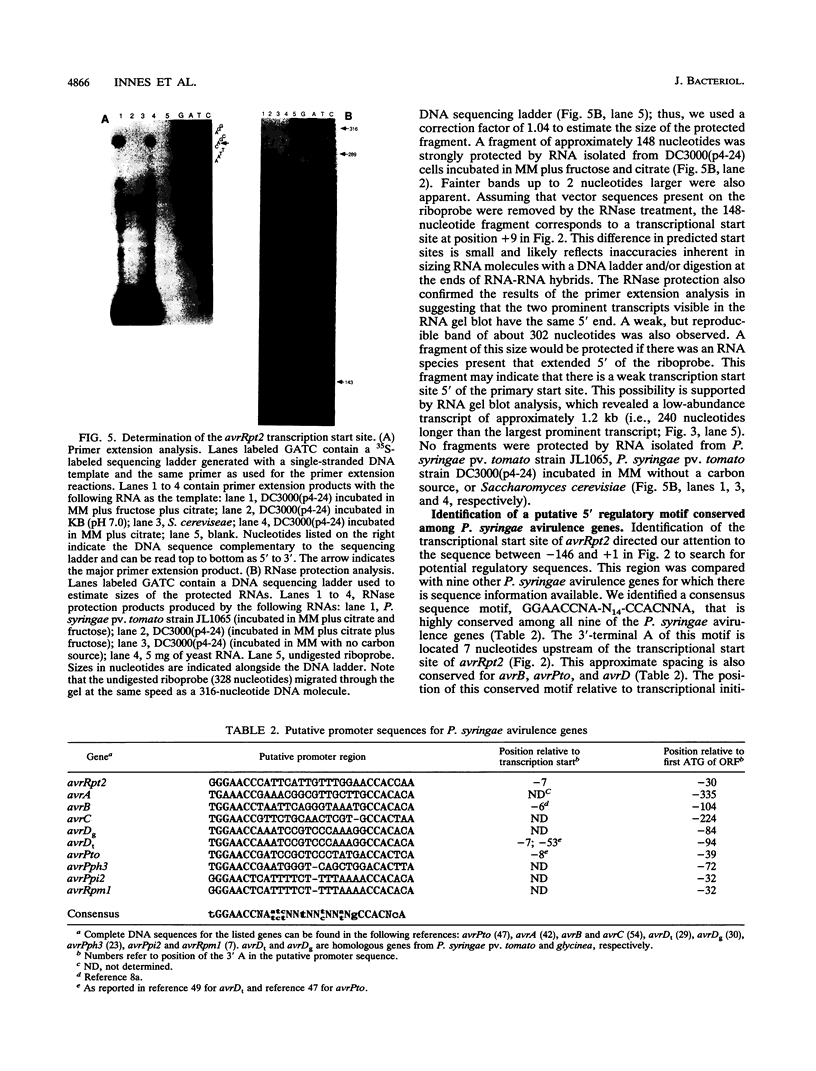
The avrRpt2 locus from Pseudomonas syringae pv. tomato causes virulent strains of P. syringae to be avirulent on some, but not all, lines of Arabidopsis thaliana and Glycine max (soybean). We determined the DNA sequence of the avrRpt2 locus and identified the avrRpt2 gene as a 768-bp open reading frame encoding a putative 28.2-kDa protein. Deletion analysis and transcription studies provided further evidence that this open reading frame encodes AvrRpt2. We found that the avrRpt2 gene also has avirulence activity in P. syringae pathogens of Phaseolus vulgaris (common bean), suggesting that disease resistance genes specific to avrRpt2 are functionally conserved among diverse plant species. The predicted AvrRpt2 protein is hydrophilic and contains no obvious membrane-spanning domains or export signal sequences, and there was no significant similarity of AvrRpt2 to sequences in the GenBank, EMBL, or Swiss PIR data bases. A comparison of the avrRpt2 DNA sequence to nine other P. syringae avirulence genes revealed a highly conserved sequence, GGAACCNA-N14-CCACNNA, upstream of the translation initiation codon. This motif is located 6 to 8 nucleotides upstream of the transcription start site in all four P. syringae avirulence genes for which a transcription start site has been determined, suggesting a role as a binding site for a novel form of RNA polymerase. Regulation of avrRpt2 was similar to other P. syringae avirulence genes; expression was high in minimal medium and low in rich medium and depended on the hrpRS locus and an additional locus at the opposite end of the hrp cluster of P. syringae pv. tomato.
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bender C. L., Cooksey D. A. Indigenous plasmids in Pseudomonas syringae pv. tomato: conjugative transfer and role in copper resistance. J Bacteriol. 1986 Feb;165(2):534–541. doi: 10.1128/jb.165.2.534-541.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown N. L., Misra T. K., Winnie J. N., Schmidt A., Seiff M., Silver S. The nucleotide sequence of the mercuric resistance operons of plasmid R100 and transposon Tn501: further evidence for mer genes which enhance the activity of the mercuric ion detoxification system. Mol Gen Genet. 1986 Jan;202(1):143–151. doi: 10.1007/BF00330531. [DOI] [PubMed] [Google Scholar]
- Collado-Vides J., Magasanik B., Gralla J. D. Control site location and transcriptional regulation in Escherichia coli. Microbiol Rev. 1991 Sep;55(3):371–394. doi: 10.1128/mr.55.3.371-394.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangl J. L., Ritter C., Gibbon M. J., Mur L. A., Wood J. R., Goss S., Mansfield J., Taylor J. D., Vivian A. Functional homologs of the Arabidopsis RPM1 disease resistance gene in bean and pea. Plant Cell. 1992 Nov;4(11):1359–1369. doi: 10.1105/tpc.4.11.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X., Mindrinos M., Davis K. R., Ausubel F. M. Induction of Arabidopsis defense genes by virulent and avirulent Pseudomonas syringae strains and by a cloned avirulence gene. Plant Cell. 1991 Jan;3(1):61–72. doi: 10.1105/tpc.3.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckhardt T. A rapid method for the identification of plasmid desoxyribonucleic acid in bacteria. Plasmid. 1978 Sep;1(4):584–588. doi: 10.1016/0147-619x(78)90016-1. [DOI] [PubMed] [Google Scholar]
- Fenselau S., Balbo I., Bonas U. Determinants of pathogenicity in Xanthomonas campestris pv. vesicatoria are related to proteins involved in secretion in bacterial pathogens of animals. Mol Plant Microbe Interact. 1992 Sep-Oct;5(5):390–396. doi: 10.1094/mpmi-5-390. [DOI] [PubMed] [Google Scholar]
- Figurski D. H., Helinski D. R. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gough C. L., Genin S., Zischek C., Boucher C. A. hrp genes of Pseudomonas solanacearum are homologous to pathogenicity determinants of animal pathogenic bacteria and are conserved among plant pathogenic bacteria. Mol Plant Microbe Interact. 1992 Sep-Oct;5(5):384–389. doi: 10.1094/mpmi-5-384. [DOI] [PubMed] [Google Scholar]
- Grimm C., Panopoulos N. J. The predicted protein product of a pathogenicity locus from Pseudomonas syringae pv. phaseolicola is homologous to a highly conserved domain of several procaryotic regulatory proteins. J Bacteriol. 1989 Sep;171(9):5031–5038. doi: 10.1128/jb.171.9.5031-5038.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H. C., He S. Y., Bauer D. W., Collmer A. The Pseudomonas syringae pv. syringae 61 hrpH product, an envelope protein required for elicitation of the hypersensitive response in plants. J Bacteriol. 1992 Nov;174(21):6878–6885. doi: 10.1128/jb.174.21.6878-6885.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh T. V., Dahlbeck D., Staskawicz B. J. Bacterial blight of soybean: regulation of a pathogen gene determining host cultivar specificity. Science. 1989 Sep 22;245(4924):1374–1377. doi: 10.1126/science.2781284. [DOI] [PubMed] [Google Scholar]
- Inouye S., Ebina Y., Nakazawa A., Nakazawa T. Nucleotide sequence surrounding transcription initiation site of xylABC operon on TOL plasmid of Pseudomonas putida. Proc Natl Acad Sci U S A. 1984 Mar;81(6):1688–1691. doi: 10.1073/pnas.81.6.1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenner C., Hitchin E., Mansfield J., Walters K., Betteridge P., Teverson D., Taylor J. Gene-for-gene interactions between Pseudomonas syringae pv. phaseolicola and Phaseolus. Mol Plant Microbe Interact. 1991 Nov-Dec;4(6):553–562. [PubMed] [Google Scholar]
- KING E. O., WARD M. K., RANEY D. E. Two simple media for the demonstration of pyocyanin and fluorescin. J Lab Clin Med. 1954 Aug;44(2):301–307. [PubMed] [Google Scholar]
- Keen N. T. Gene-for-gene complementarity in plant-pathogen interactions. Annu Rev Genet. 1990;24:447–463. doi: 10.1146/annurev.ge.24.120190.002311. [DOI] [PubMed] [Google Scholar]
- Keen N. T. The molecular biology of disease resistance. Plant Mol Biol. 1992 May;19(1):109–122. doi: 10.1007/BF00015609. [DOI] [PubMed] [Google Scholar]
- Kobayashi D. Y., Tamaki S. J., Keen N. T. Cloned avirulence genes from the tomato pathogen Pseudomonas syringae pv. tomato confer cultivar specificity on soybean. Proc Natl Acad Sci U S A. 1989 Jan;86(1):157–161. doi: 10.1073/pnas.86.1.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi D. Y., Tamaki S. J., Keen N. T. Molecular characterization of avirulence gene D from Pseudomonas syringae pv. tomato. Mol Plant Microbe Interact. 1990 Mar-Apr;3(2):94–102. doi: 10.1094/mpmi-3-094. [DOI] [PubMed] [Google Scholar]
- Kobayashi D. Y., Tamaki S. J., Trollinger D. J., Gold S., Keen N. T. A gene from Pseudomonas syringae pv. glycinea with homology to avirulence gene D from P. s. pv. tomato but devoid of the avirulence phenotype. Mol Plant Microbe Interact. 1990 Mar-Apr;3(2):103–111. doi: 10.1094/mpmi-3-103. [DOI] [PubMed] [Google Scholar]
- Kustu S., Santero E., Keener J., Popham D., Weiss D. Expression of sigma 54 (ntrA)-dependent genes is probably united by a common mechanism. Microbiol Rev. 1989 Sep;53(3):367–376. doi: 10.1128/mr.53.3.367-376.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Lindgren P. B., Peet R. C., Panopoulos N. J. Gene cluster of Pseudomonas syringae pv. "phaseolicola" controls pathogenicity of bean plants and hypersensitivity of nonhost plants. J Bacteriol. 1986 Nov;168(2):512–522. doi: 10.1128/jb.168.2.512-522.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lory S. Determinants of extracellular protein secretion in gram-negative bacteria. J Bacteriol. 1992 Jun;174(11):3423–3428. doi: 10.1128/jb.174.11.3423-3428.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michiels T., Cornelis G. R. Secretion of hybrid proteins by the Yersinia Yop export system. J Bacteriol. 1991 Mar;173(5):1677–1685. doi: 10.1128/jb.173.5.1677-1685.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minton N. P., Clarke L. E. Identification of the promoter of the Pseudomonas gene coding for carboxypeptidase G2. J Mol Appl Genet. 1985;3(1):26–35. [PubMed] [Google Scholar]
- Mott J. E., Galloway J. L., Platt T. Maturation of Escherichia coli tryptophan operon mRNA: evidence for 3' exonucleolytic processing after rho-dependent termination. EMBO J. 1985 Jul;4(7):1887–1891. doi: 10.1002/j.1460-2075.1985.tb03865.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napoli C., Staskawicz B. Molecular characterization and nucleic acid sequence of an avirulence gene from race 6 of Pseudomonas syringae pv. glycinea. J Bacteriol. 1987 Feb;169(2):572–578. doi: 10.1128/jb.169.2.572-578.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prentki P., Krisch H. M. In vitro insertional mutagenesis with a selectable DNA fragment. Gene. 1984 Sep;29(3):303–313. doi: 10.1016/0378-1119(84)90059-3. [DOI] [PubMed] [Google Scholar]
- Rahme L. G., Mindrinos M. N., Panopoulos N. J. Genetic and transcriptional organization of the hrp cluster of Pseudomonas syringae pv. phaseolicola. J Bacteriol. 1991 Jan;173(2):575–586. doi: 10.1128/jb.173.2.575-586.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahme L. G., Mindrinos M. N., Panopoulos N. J. Plant and environmental sensory signals control the expression of hrp genes in Pseudomonas syringae pv. phaseolicola. J Bacteriol. 1992 Jun;174(11):3499–3507. doi: 10.1128/jb.174.11.3499-3507.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos J. L., Mermod N., Timmis K. N. Regulatory circuits controlling transcription of TOL plasmid operon encoding meta-cleavage pathway for degradation of alkylbenzoates by Pseudomonas. Mol Microbiol. 1987 Nov;1(3):293–300. doi: 10.1111/j.1365-2958.1987.tb01935.x. [DOI] [PubMed] [Google Scholar]
- Salmeron J. M., Staskawicz B. J. Molecular characterization and hrp dependence of the avirulence gene avrPto from Pseudomonas syringae pv. tomato [corrected]. Mol Gen Genet. 1993 May;239(1-2):6–16. doi: 10.1007/BF00281595. [DOI] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. The 3'-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staskawicz B., Dahlbeck D., Keen N., Napoli C. Molecular characterization of cloned avirulence genes from race 0 and race 1 of Pseudomonas syringae pv. glycinea. J Bacteriol. 1987 Dec;169(12):5789–5794. doi: 10.1128/jb.169.12.5789-5794.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaki S., Dahlbeck D., Staskawicz B., Keen N. T. Characterization and expression of two avirulence genes cloned from Pseudomonas syringae pv. glycinea. J Bacteriol. 1988 Oct;170(10):4846–4854. doi: 10.1128/jb.170.10.4846-4854.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J., Messing J. Production of single-stranded plasmid DNA. Methods Enzymol. 1987;153:3–11. doi: 10.1016/0076-6879(87)53044-0. [DOI] [PubMed] [Google Scholar]
- Whalen M. C., Innes R. W., Bent A. F., Staskawicz B. J. Identification of Pseudomonas syringae pathogens of Arabidopsis and a bacterial locus determining avirulence on both Arabidopsis and soybean. Plant Cell. 1991 Jan;3(1):49–59. doi: 10.1105/tpc.3.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whalen M. C., Stall R. E., Staskawicz B. J. Characterization of a gene from a tomato pathogen determining hypersensitive resistance in non-host species and genetic analysis of this resistance in bean. Proc Natl Acad Sci U S A. 1988 Sep;85(18):6743–6747. doi: 10.1073/pnas.85.18.6743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y., Lu Y., Heu S., Hutcheson S. W. Organization and environmental regulation of the Pseudomonas syringae pv. syringae 61 hrp cluster. J Bacteriol. 1992 Mar;174(6):1734–1741. doi: 10.1128/jb.174.6.1734-1741.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]





