Abstract
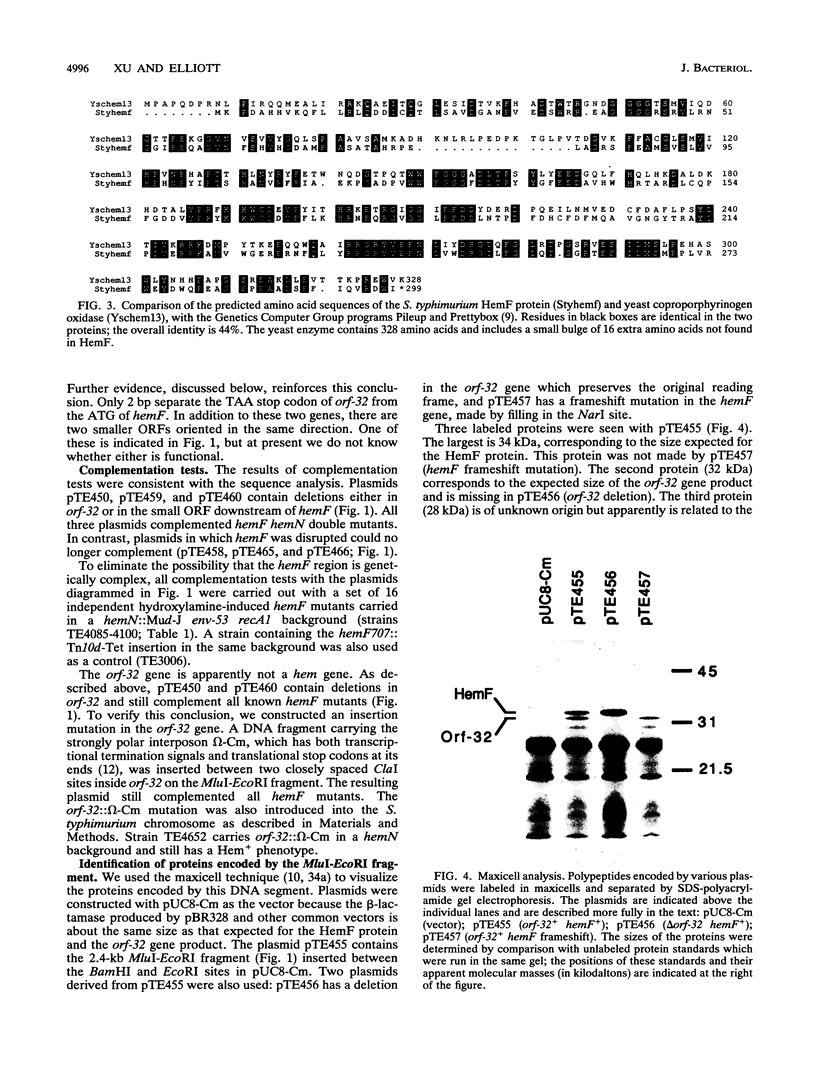
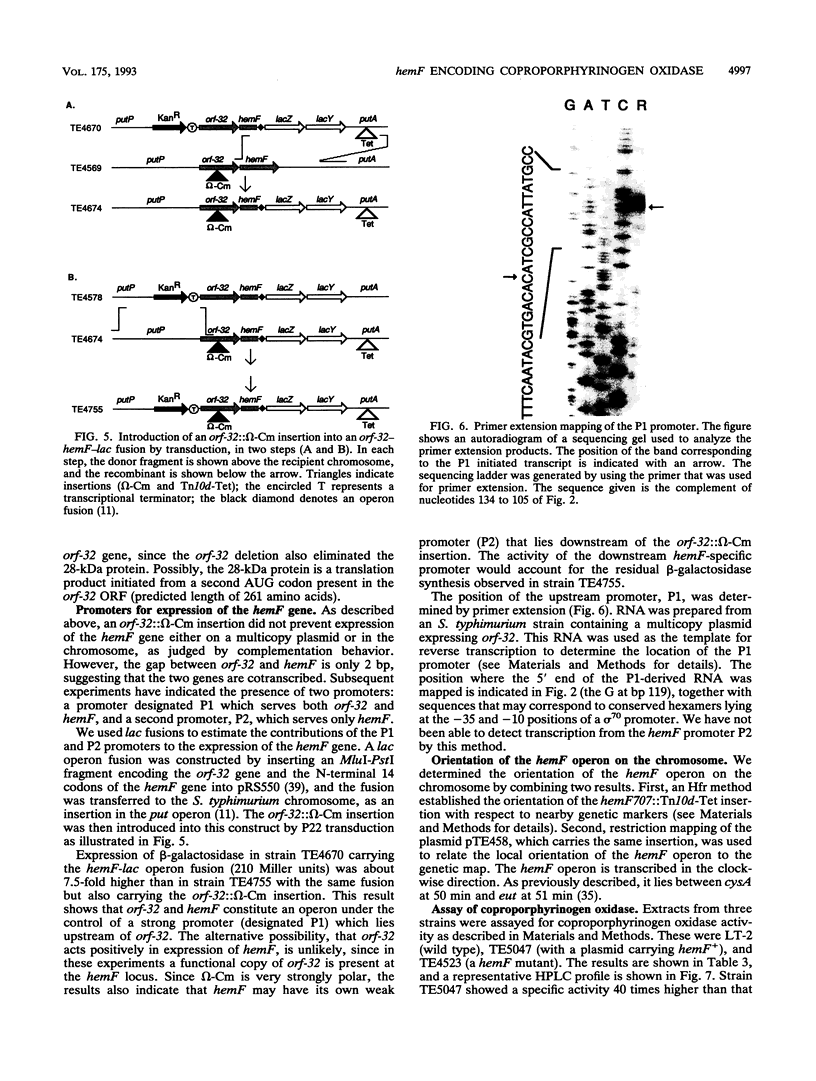
The 8th step in the 10-step heme biosynthetic pathway of Salmonella typhimurium is the oxidation of coproporphyrinogen III to protoporphyrinogen IX. On the basis of genetic studies, we have suggested that this reaction may be catalyzed by either of two different enzymes, an oxygen-dependent one encoded by hemF or an oxygen-independent enzyme encoded by hemN. Here, we report the cloning of the S. typhimurium hemF gene and its DNA sequence. The predicted amino acid sequence of the HemF protein is 44% identical to that of the coproporphyrinogen oxidase encoded by the yeast HEM13 gene. The wild-type S. typhimurium strain LT-2 produces an oxygen-dependent coproporphyrinogen oxidase activity detectable in crude extracts, which is not found in hemF mutants and is overproduced in strains carrying the hemF gene on a multicopy plasmid. the hemF gene is the second gene in an operon with an upstream gene with an unknown function, whose amino acid sequence suggests a relation to amidases involved in cell wall synthesis or remodeling. The upstream gene and hemF are cotranscribed from a promoter which was mapped by primer extension. A weaker, hemF-specific promoter is inferred from the behavior of an omega-Cm insertion mutation in the upstream gene. Although this insertion decreases expression of beta-galactosidase about 7.5-fold when placed upstream of a hemF-lacZ operon fusion, it still allows sufficient HemF expression from an otherwise wild-type construct to confer a Hem+ phenotype. The hemF operon is transcribed clockwise with respect to the genetic map.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berkowitz D., Hushon J. M., Whitfield H. J., Jr, Roth J., Ames B. N. Procedure for identifying nonsense mutations. J Bacteriol. 1968 Jul;96(1):215–220. doi: 10.1128/jb.96.1.215-220.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boorstein W. R., Craig E. A. Primer extension analysis of RNA. Methods Enzymol. 1989;180:347–369. doi: 10.1016/0076-6879(89)80111-9. [DOI] [PubMed] [Google Scholar]
- Chen E. Y., Seeburg P. H. Supercoil sequencing: a fast and simple method for sequencing plasmid DNA. DNA. 1985 Apr;4(2):165–170. doi: 10.1089/dna.1985.4.165. [DOI] [PubMed] [Google Scholar]
- Chumley F. G., Menzel R., Roth J. R. Hfr formation directed by tn10. Genetics. 1979 Apr;91(4):639–655. doi: 10.1093/genetics/91.4.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott T. A method for constructing single-copy lac fusions in Salmonella typhimurium and its application to the hemA-prfA operon. J Bacteriol. 1992 Jan;174(1):245–253. doi: 10.1128/jb.174.1.245-253.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott T. Cloning, genetic characterization, and nucleotide sequence of the hemA-prfA operon of Salmonella typhimurium. J Bacteriol. 1989 Jul;171(7):3948–3960. doi: 10.1128/jb.171.7.3948-3960.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fellay R., Frey J., Krisch H. Interposon mutagenesis of soil and water bacteria: a family of DNA fragments designed for in vitro insertional mutagenesis of gram-negative bacteria. Gene. 1987;52(2-3):147–154. doi: 10.1016/0378-1119(87)90041-2. [DOI] [PubMed] [Google Scholar]
- Foster T. J., Davis M. A., Roberts D. E., Takeshita K., Kleckner N. Genetic organization of transposon Tn10. Cell. 1981 Jan;23(1):201–213. doi: 10.1016/0092-8674(81)90285-3. [DOI] [PubMed] [Google Scholar]
- Hahn D. R., Myers R. S., Kent C. R., Maloy S. R. Regulation of proline utilization in Salmonella typhimurium: molecular characterization of the put operon, and DNA sequence of the put control region. Mol Gen Genet. 1988 Jul;213(1):125–133. doi: 10.1007/BF00333408. [DOI] [PubMed] [Google Scholar]
- Hall M. N., Hereford L., Herskowitz I. Targeting of E. coli beta-galactosidase to the nucleus in yeast. Cell. 1984 Apr;36(4):1057–1065. doi: 10.1016/0092-8674(84)90055-2. [DOI] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984 Jun;28(3):351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- Hmiel S. P., Snavely M. D., Miller C. G., Maguire M. E. Magnesium transport in Salmonella typhimurium: characterization of magnesium influx and cloning of a transport gene. J Bacteriol. 1986 Dec;168(3):1444–1450. doi: 10.1128/jb.168.3.1444-1450.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong J. S., Ames B. N. Localized mutagenesis of any specific small region of the bacterial chromosome. Proc Natl Acad Sci U S A. 1971 Dec;68(12):3158–3162. doi: 10.1073/pnas.68.12.3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingledew W. J., Poole R. K. The respiratory chains of Escherichia coli. Microbiol Rev. 1984 Sep;48(3):222–271. doi: 10.1128/mr.48.3.222-271.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs N. J., Jacobs J. M. Assay for enzymatic protoporphyrinogen oxidation, a late step in heme synthesis. Enzyme. 1982;28(2-3):206–219. doi: 10.1159/000459103. [DOI] [PubMed] [Google Scholar]
- Janzer J. J., Stan-Lotter H., Sanderson K. E. Isolation and characterization of hemin-permeable, envelope-defective mutants of Salmonella typhimurium. Can J Microbiol. 1981 Feb;27(2):226–237. doi: 10.1139/m81-034. [DOI] [PubMed] [Google Scholar]
- Keithly J. H., Nadler K. D. Protoporphyrin formation in Rhizobium japonicum. J Bacteriol. 1983 May;154(2):838–845. doi: 10.1128/jb.154.2.838-845.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleckner N., Steele D. A., Reichardt K., Botstein D. Specificity of insertion by the translocatable tetracycline-resistance element Tn10. Genetics. 1979 Aug;92(4):1023–1040. doi: 10.1093/genetics/92.4.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda A., Sekiguchi J. Molecular cloning and sequencing of a major Bacillus subtilis autolysin gene. J Bacteriol. 1991 Nov;173(22):7304–7312. doi: 10.1128/jb.173.22.7304-7312.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Lazarevic V., Margot P., Soldo B., Karamata D. Sequencing and analysis of the Bacillus subtilis lytRABC divergon: a regulatory unit encompassing the structural genes of the N-acetylmuramoyl-L-alanine amidase and its modifier. J Gen Microbiol. 1992 Sep;138(9):1949–1961. doi: 10.1099/00221287-138-9-1949. [DOI] [PubMed] [Google Scholar]
- Mankovich J. A., McIntyre C. A., Walker G. C. Nucleotide sequence of the Salmonella typhimurium mutL gene required for mismatch repair: homology of MutL to HexB of Streptococcus pneumoniae and to PMS1 of the yeast Saccharomyces cerevisiae. J Bacteriol. 1989 Oct;171(10):5325–5331. doi: 10.1128/jb.171.10.5325-5331.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porra R. J., Falk J. E. The enzymic conversion of coproporphyrinogen 3 into protoporphyrin 9. Biochem J. 1964 Jan;90(1):69–75. doi: 10.1042/bj0900069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roland K. L., Powell F. E., Turnbough C. L., Jr Role of translation and attenuation in the control of pyrBI operon expression in Escherichia coli K-12. J Bacteriol. 1985 Sep;163(3):991–999. doi: 10.1128/jb.163.3.991-999.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SANO S., GRANICK S. Mitochondrial coproporphyrinogen oxidase and protoporphyrin formation. J Biol Chem. 1961 Apr;236:1173–1180. [PubMed] [Google Scholar]
- Sancar A., Hack A. M., Rupp W. D. Simple method for identification of plasmid-coded proteins. J Bacteriol. 1979 Jan;137(1):692–693. doi: 10.1128/jb.137.1.692-693.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson K. E., Roth J. R. Linkage map of Salmonella typhimurium, edition VII. Microbiol Rev. 1988 Dec;52(4):485–532. doi: 10.1128/mr.52.4.485-532.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmieger H. Phage P22-mutants with increased or decreased transduction abilities. Mol Gen Genet. 1972;119(1):75–88. doi: 10.1007/BF00270447. [DOI] [PubMed] [Google Scholar]
- Seehra J. S., Jordan P. M., Akhtar M. Anaerobic and aerobic coproporphyrinogen III oxidases of Rhodopseudomonas spheroides. Mechanism and stereochemistry of vinyl group formation. Biochem J. 1983 Mar 1;209(3):709–718. doi: 10.1042/bj2090709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons R. W., Houman F., Kleckner N. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene. 1987;53(1):85–96. doi: 10.1016/0378-1119(87)90095-3. [DOI] [PubMed] [Google Scholar]
- Straka J. G. High-performance liquid chromatography of porphyrin methyl esters. Methods Enzymol. 1986;123:352–363. doi: 10.1016/s0076-6879(86)23042-6. [DOI] [PubMed] [Google Scholar]
- Tait G. H. Coproporphyrinogenase activities in extracts of Rhodopseudomonas spheroides and Chromatium strain D. Biochem J. 1972 Aug;128(5):1159–1169. doi: 10.1042/bj1281159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomioka S., Nikaido T., Miyakawa T., Matsuhashi M. Mutation of the N-acetylmuramyl-L-alanine amidase gene of Escherichia coli K-12. J Bacteriol. 1983 Oct;156(1):463–465. doi: 10.1128/jb.156.1.463-465.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J., Messing J. Production of single-stranded plasmid DNA. Methods Enzymol. 1987;153:3–11. doi: 10.1016/0076-6879(87)53044-0. [DOI] [PubMed] [Google Scholar]
- Whitfield H. J., Levine G. Isolation and characterization of a mutant of Salmonella typhimurium deficient in a major deoxyribonucleic acid polymerase activity. J Bacteriol. 1973 Oct;116(1):54–58. doi: 10.1128/jb.116.1.54-58.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson H. R., Archer C. D., Liu J. K., Turnbough C. L., Jr Translational control of pyrC expression mediated by nucleotide-sensitive selection of transcriptional start sites in Escherichia coli. J Bacteriol. 1992 Jan;174(2):514–524. doi: 10.1128/jb.174.2.514-524.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K., Delling J., Elliott T. The genes required for heme synthesis in Salmonella typhimurium include those encoding alternative functions for aerobic and anaerobic coproporphyrinogen oxidation. J Bacteriol. 1992 Jun;174(12):3953–3963. doi: 10.1128/jb.174.12.3953-3963.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zagorec M., Buhler J. M., Treich I., Keng T., Guarente L., Labbe-Bois R. Isolation, sequence, and regulation by oxygen of the yeast HEM13 gene coding for coproporphyrinogen oxidase. J Biol Chem. 1988 Jul 15;263(20):9718–9724. [PubMed] [Google Scholar]




