Abstract
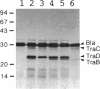
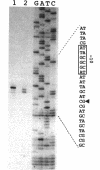
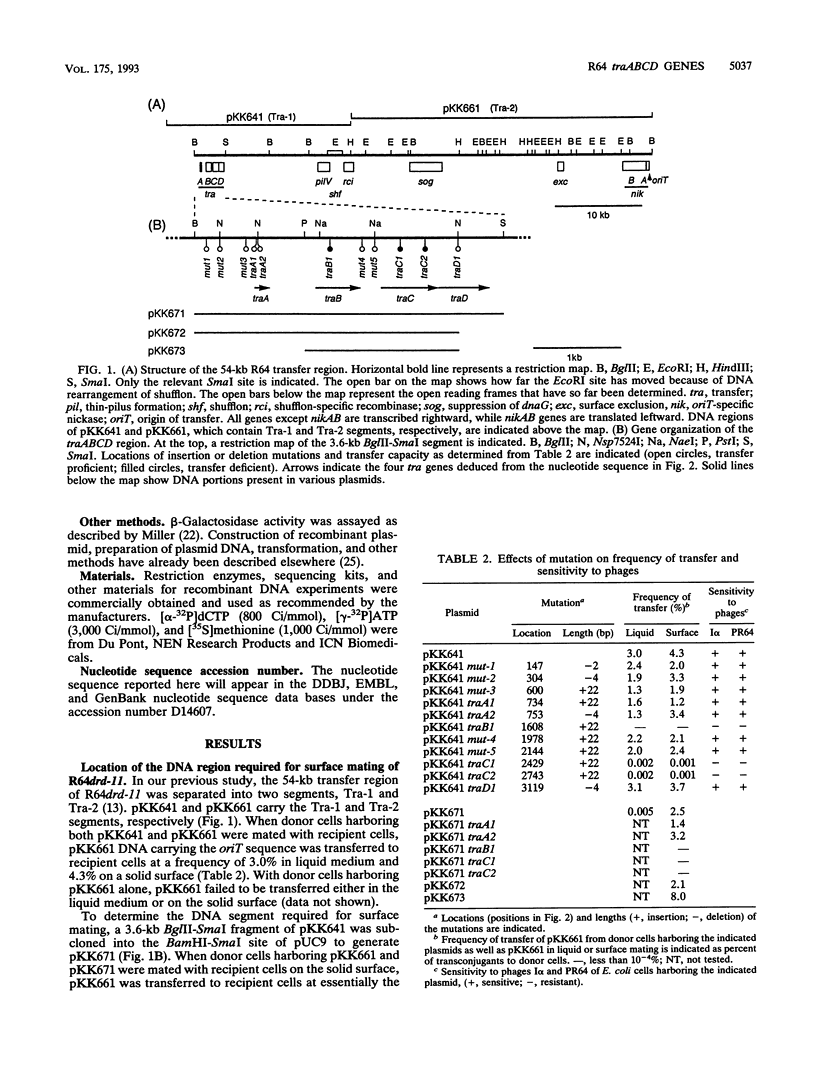
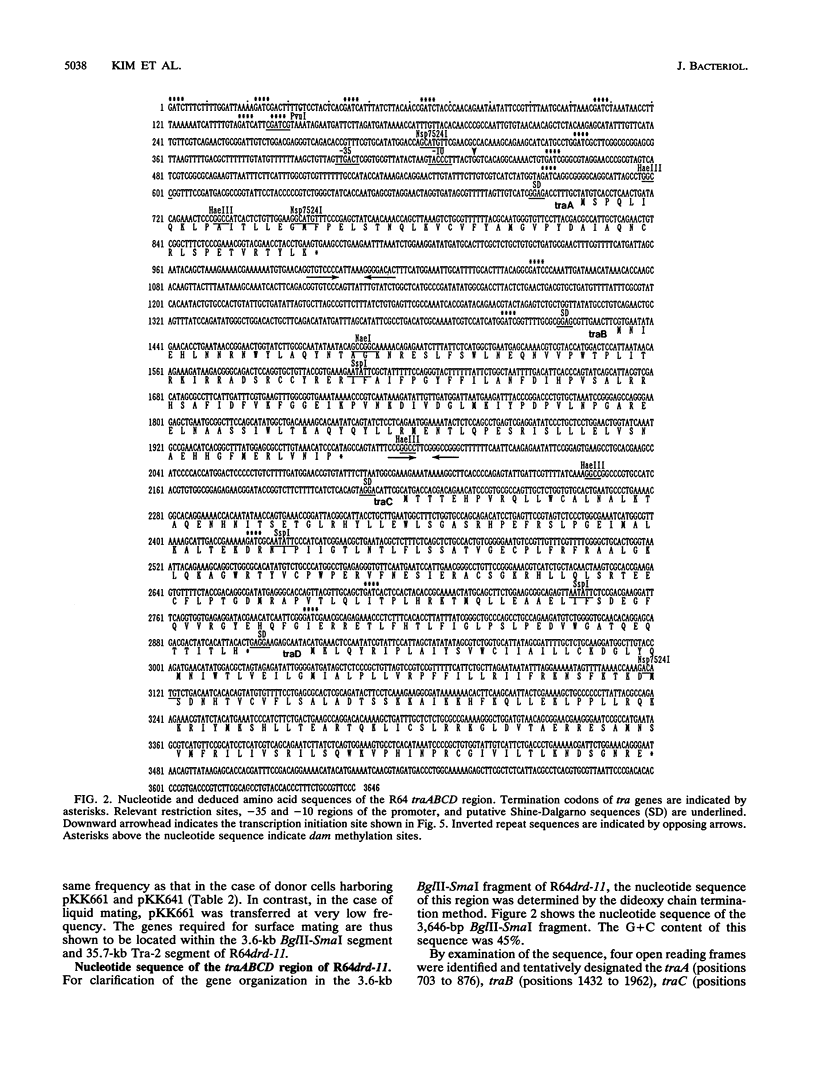
A 3.6-kb BglII-SmaI segment of the transfer region of IncI1 plasmid R64drd-11 was sequenced and characterized. Analysis of the DNA sequence indicated the presence of four genes, traA, traB, traC, and traD, in this region. The expression of the traB, traC, and traD genes was examined by maxicell experiments and that of the traA gene was examined by constructing the traA-lacZ fusion gene. The introduction of frameshift mutations into the four genes indicated that the traB and traC genes are essential for conjugal transfer in liquid medium and on a solid surface. Both were also required for the formation of the thin pilus, which is the receptor for phages I alpha and PR64FS. Upstream of the traA gene, a promoter sequence for sigma 70 of E. coli RNA polymerase was identified by S1 nuclease mapping and primer extension experiments.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aiba H., Adhya S., de Crombrugghe B. Evidence for two functional gal promoters in intact Escherichia coli cells. J Biol Chem. 1981 Nov 25;256(22):11905–11910. [PubMed] [Google Scholar]
- Bradley D. E. Characteristics and function of thick and thin conjugative pili determined by transfer-derepressed plasmids of incompatibility groups I1, I2, I5, B, K and Z. J Gen Microbiol. 1984 Jun;130(6):1489–1502. doi: 10.1099/00221287-130-6-1489. [DOI] [PubMed] [Google Scholar]
- Casadaban M. J., Chou J., Cohen S. N. In vitro gene fusions that join an enzymatically active beta-galactosidase segment to amino-terminal fragments of exogenous proteins: Escherichia coli plasmid vectors for the detection and cloning of translational initiation signals. J Bacteriol. 1980 Aug;143(2):971–980. doi: 10.1128/jb.143.2.971-980.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coetzee J. N., Bradley D. E., Hedges R. W. Phages I alpha and I2-2: IncI plasmid-dependent bacteriophages. J Gen Microbiol. 1982 Nov;128(11):2797–2804. doi: 10.1099/00221287-128-11-2797. [DOI] [PubMed] [Google Scholar]
- Coetzee J. N., Sirgel F. A., Lecatsas G. Properties of a filamentous phage which adsorbs to pili coded by plasmids of the IncI complex. J Gen Microbiol. 1980 Apr;117(2):547–551. doi: 10.1099/00221287-117-2-547. [DOI] [PubMed] [Google Scholar]
- Frost L., Lee S., Yanchar N., Paranchych W. finP and fisO mutations in FinP anti-sense RNA suggest a model for FinOP action in the repression of bacterial conjugation by the Flac plasmid JCFL0. Mol Gen Genet. 1989 Jul;218(1):152–160. doi: 10.1007/BF00330578. [DOI] [PubMed] [Google Scholar]
- Furuya N., Nisioka T., Komano T. Nucleotide sequence and functions of the oriT operon in IncI1 plasmid R64. J Bacteriol. 1991 Apr;173(7):2231–2237. doi: 10.1128/jb.173.7.2231-2237.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harley C. B., Reynolds R. P. Analysis of E. coli promoter sequences. Nucleic Acids Res. 1987 Mar 11;15(5):2343–2361. doi: 10.1093/nar/15.5.2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartskeerl R., Overduin P., Hoekstra W., Tommassen J. Nucleotide sequence of the exclusion-determining locus of IncI plasmid R144. Gene. 1986;42(1):107–111. doi: 10.1016/0378-1119(86)90156-3. [DOI] [PubMed] [Google Scholar]
- Komano T., Funayama N., Kim S. R., Nisioka T. Transfer region of IncI1 plasmid R64 and role of shufflon in R64 transfer. J Bacteriol. 1990 May;172(5):2230–2235. doi: 10.1128/jb.172.5.2230-2235.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komano T., Kim S. R., Nisioka T. Distribution of shufflon among IncI plasmids. J Bacteriol. 1987 Nov;169(11):5317–5319. doi: 10.1128/jb.169.11.5317-5319.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komano T., Kubo A., Kayanuma T., Furuichi T., Nisioka T. Highly mobile DNA segment of IncI alpha plasmid R64: a clustered inversion region. J Bacteriol. 1986 Jan;165(1):94–100. doi: 10.1128/jb.165.1.94-100.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komano T., Kubo A., Nisioka T. Shufflon: multi-inversion of four contiguous DNA segments of plasmid R64 creates seven different open reading frames. Nucleic Acids Res. 1987 Feb 11;15(3):1165–1172. doi: 10.1093/nar/15.3.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo A., Kusukawa A., Komano T. Nucleotide sequence of the rci gene encoding shufflon-specific DNA recombinase in the IncI1 plasmid R64: homology to the site-specific recombinases of integrase family. Mol Gen Genet. 1988 Jul;213(1):30–35. doi: 10.1007/BF00333394. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K., Favre M. Maturation of the head of bacteriophage T4. I. DNA packaging events. J Mol Biol. 1973 Nov 15;80(4):575–599. doi: 10.1016/0022-2836(73)90198-8. [DOI] [PubMed] [Google Scholar]
- Marinus M. G., Morris N. R. Biological function for 6-methyladenine residues in the DNA of Escherichia coli K12. J Mol Biol. 1974 May 15;85(2):309–322. doi: 10.1016/0022-2836(74)90366-0. [DOI] [PubMed] [Google Scholar]
- Merryweather A., Rees C. E., Smith N. M., Wilkins B. M. Role of sog polypeptides specified by plasmid ColIb-P9 and their transfer between conjugating bacteria. EMBO J. 1986 Nov;5(11):3007–3012. doi: 10.1002/j.1460-2075.1986.tb04599.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messer W., Bellekes U., Lother H. Effect of dam methylation on the activity of the E. coli replication origin, oriC. EMBO J. 1985 May;4(5):1327–1332. doi: 10.1002/j.1460-2075.1985.tb03780.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees C. E., Bradley D. E., Wilkins B. M. Organization and regulation of the conjugation genes of IncI1 plasmid colIb-P9. Plasmid. 1987 Nov;18(3):223–236. doi: 10.1016/0147-619x(87)90065-5. [DOI] [PubMed] [Google Scholar]
- Roberts D., Hoopes B. C., McClure W. R., Kleckner N. IS10 transposition is regulated by DNA adenine methylation. Cell. 1985 Nov;43(1):117–130. doi: 10.1016/0092-8674(85)90017-0. [DOI] [PubMed] [Google Scholar]
- Sancar A., Hack A. M., Rupp W. D. Simple method for identification of plasmid-coded proteins. J Bacteriol. 1979 Jan;137(1):692–693. doi: 10.1128/jb.137.1.692-693.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D. W., Garland A. M., Herman G., Enns R. E., Baker T. A., Zyskind J. W. Importance of state of methylation of oriC GATC sites in initiation of DNA replication in Escherichia coli. EMBO J. 1985 May;4(5):1319–1326. doi: 10.1002/j.1460-2075.1985.tb03779.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeshita S., Sato M., Toba M., Masahashi W., Hashimoto-Gotoh T. High-copy-number and low-copy-number plasmid vectors for lacZ alpha-complementation and chloramphenicol- or kanamycin-resistance selection. Gene. 1987;61(1):63–74. doi: 10.1016/0378-1119(87)90365-9. [DOI] [PubMed] [Google Scholar]
- Thompson R., Taylor L. Promoter mapping and DNA sequencing of the F plasmid transfer genes traM and traJ. Mol Gen Genet. 1982;188(3):513–518. doi: 10.1007/BF00330058. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Yoshida T., Takahashi I., Tubahara H., Sasakawa C., Yoshikawa M. Significance of filter mating in integrative incompatibility test for plasmid classification. Microbiol Immunol. 1984;28(1):63–73. doi: 10.1111/j.1348-0421.1984.tb02947.x. [DOI] [PubMed] [Google Scholar]