Abstract
Regulation of gene expression by many transcription factors is controlled by specific combinations of homo- and heterodimers through a short α-helical coiled-coil known as a leucine zipper. The dimer interface of a leucine zipper involves side chains of the residues at the a, d, e, and g positions of the (abcdefg)n heptad repeat. To understand the basis for the specificity of dimer formation, we characterized GCN4 leucine zipper mutants with all 16 possible permutations and combinations of isoleucines and asparagines at four a positions in the dimer interface, using a genetic test for the specificity of dimer formation by λ repressor-leucine zipper fusions. Heterodimers were detected by loss of repressor activity in the presence of a fusion to a dominant-negative mutant form of the DNA-binding domain of repressor. Reconstruction experiments using leucine zippers from GCN4, Jun, Fos, and C/EBP showed that this assay distinguishes pairs that form heterodimers from those that do not. We found that the mutants have novel dimerization specificities determined by the positioning of buried asparagine residues at the a positions. The pattern of buried polar residues could also explain the dimerization specificities of some naturally occurring leucine zippers. The altered specificity mutants described here should be useful for the construction of artificial regulatory circuitry.
Keywords: protein structure, site-directed mutagenesis, recombinant fusion proteins, genetics
The stoichiometry and specificity with which proteins interact is a key control point in many biological processes. For example, common dimerization domains allow transcription factors in the bZIP or bHLH-LZ families to form a variety of homo- and heterodimers with different properties. By expressing different sets of subunits under different conditions, cells can generate complex regulatory circuits from a relatively small number of genes. The correct functioning of this complex regulatory machinery depends on each of the component proteins assembling only with specific partners.
Leucine zippers are an excellent model system to study how the stability and specificity of protein–protein interactions are determined. High-resolution x-ray crystallographic and NMR structures are available for several leucine zippers (1–7). As α-helical coiled coils, leucine zippers have simple secondary and tertiary structures. Moreover, the large number of naturally occurring leucine zipper proteins includes a wide variety of distinct and overlapping dimerization specificities.
At the amino acid sequence level, leucine zippers are characterized by leucine appearing in every seventh position (d) over 4 to 5 heptad repeats (abcdefg)n. The hydrophobic core of the dimer interface is formed by residues at the a and d positions (Fig. 1); the solvent-accessible e and g positions are frequently occupied by charged amino acids (8, 9). In the crystal structures of leucine zippers, including GCN4 homodimers and Jun–Fos heterodimers, intersubunit salt bridges are seen between oppositely charged amino acids at the g (ith heptad) and e′ (i + 1th heptad of the other monomer) positions (1, 2, 5, 10).
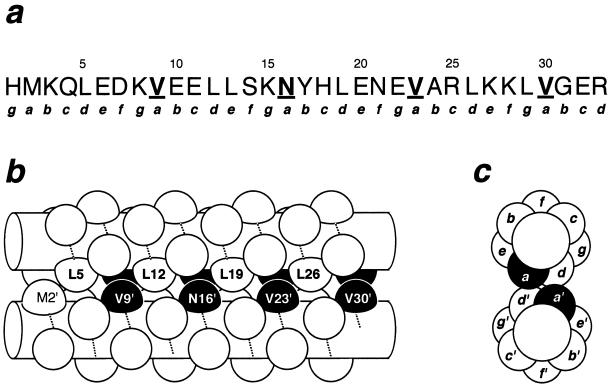
Figure 1.
The leucine zipper of GCN4 arranged as a parallel coiled coil. (a) Sequence of the leucine zipper as it occurs in the λ repressor fusion system. Lowercase letters indicate positions in the heptad repeat. The a positions targeted for mutagenesis are highlighted in boldface and are underlined. (b) Side view of the dimer. The amino acid backbones in a helical conformation are represented by cylinders with the path of the polypeptide chain indicated by the dotted lines. Side chains are represented as knobs. Residues targeted for mutagenesis are highlighted in black. (c) End view showing how different heptad positions are arranged in the dimer. The side chains at the a positions are buried in the dimer interface.
Studies on the dimerization specificity of leucine zippers have focused on these g–e′ electrostatic interactions. Mutant and engineered leucine zippers have been constructed to examine the roles of the e and g positions in dimerization specificity (11–15). These studies show that the identities of the residues at the e and g positions can be sufficient to determine the specificity of dimerization.
Other positions in the dimer interface are also important in determining the structures of leucine zippers. In the leucine zipper of the yeast transcription factor GCN4, the four d positions in each monomer are all leucines, and four of the five a positions are hydrophobic amino acids. The third a position is occupied by an asparagine at residue 16. In homodimers, the two asparagine side chains form an intersubunit hydrogen bond across the dimer interface (1, 2, 10). Both Asn side chains at position 16 of the GCN4 leucine zipper are >97% buried compared with G-X-G reference peptides, and the side-chain amides of the Asn residues at an internal a position in the Jun leucine zipper are protected from hydrogen exchange (7), indicating that they are involved in hydrogen bonds. Although this interaction is energetically less favorable than the hydrophobic and packing interactions from a pair of valine or isoleucine side chains (3, 16), the buried polar groups serve to specify the formation of dimers by destabilizing higher-order oligomers. Changing Asn-16 to Val in GCN4 leads to formation of mixtures of dimers and trimers (3, 16).
The importance of buried polar interactions in imparting structural uniqueness has been observed in other coiled coils. Designed coiled coils with entirely hydrophobic residues at the a and d positions have been found to form either mixed or unexpected oligomeric states (17, 18). An Asn-to-Leu mutation at the only polar a position in a Jun peptide leads to formation of higher-order oligomers (7). An Asn at position 14 of the ACID-p1 and BASE-p1 peptides determines formation of dimers; when this residue is mutated to Leu, heterotetramers without unique helix orientations form (19).
The hydrogen bonding of buried asparagines at the a positions and the loss of structural specificity in the absence of a buried polar group suggest that dimerization specificity will be affected by the alignment of asparagines across the dimer interface. Here, we use a genetic approach based on λ repressor fusion proteins to test this hypothesis. We find that homodimeric leucine zippers can form with asparagines at different combinations of a positions and that the positioning of buried asparagines can be sufficient to determine the specificities with which homodimeric and heterodimeric leucine zippers form.
MATERIALS AND METHODS
Microbiological Methods.
All experiments were performed on LB plates (20) in Escherichia coli strain AG1688 [F′128 lacIQ lacZ::Tn5/araD139, Δ(ara–leu)7697, Δ(lac)X74, galE15, galK16, rpsL (StrR), hsdR2, mcrA, mcrB1] (21). Plasmids were introduced into AG1688 by electroporation or M13-mediated transduction (22). Ampicillin (200 μg/ml) and tetracycline (20 μg/ml) were added to media as appropriate. Plasmid-containing cells were tested for immunity by cross-streaking against λKH54 at 37°C (21, 23).
Plasmid Vectors.
Repressor fusions to leucine zipper sequences were cloned in three different plasmid backgrounds. For “high-level” expression, leucine zippers were cloned between the SalI and BamHI sites of pJH391 (21), a pBR322-derived plasmid that expresses fusion proteins from PlacUV5. For “low-level” expression, fusion proteins were cloned in pXZ240, which is the same as pJH391 with the promoter region replaced by P7107, a mutant promoter derived from an operatorless PlacUV5 (X.Z., H. Hunter, M. Watts, and J.H., unpublished work). For C/EBP, the zipper cassette was inserted into a repressor fusion vector with a different promoter, P7051 (X.Z., H. Hunter, M. Watts, and J.H., unpublished work). cI-C/EBP does not confer immunity when expressed from P7107. However, cI+-C/EBP was able to confer immunity when expressed from P7051.
For specificity assays, fragments encoding the leucine zippers were inserted into pXZ270, which is derived from pJH550 (24), a pACYC184-based plasmid that expresses repressor fusions from the tac promoter. In pXZ270, the repressor domain has been mutated to contain a glutamine-to-leucine mutation in the DNA-recognition helix of the repressor (QL44).
Leucine Zippers.
DNA cassettes encoding the leucine zippers of GCN4, Fos, and Jun were previously described (23, 25). Synthetic DNA encoding a histidine-tagged version of the C/EBP leucine zipper was constructed by mutually primed DNA synthesis. Amino acid sequences of control leucine zippers used were as follows: GCN4, H MKQLEDK VEELLSK NYHLENE VARLKKL VGER; C/EBP, R NVETQQK VLELTSD NDRLRKR VEQLSRE LDTLR GGHHHHHH; Jun, HMRR IARLEEK VKTLKAQ NSELAST ANMLREQ VAQLKQK Y; and Fos, L TDTLQAE TDQLEDE KSALQTE IANLLKE KEKLEFI LAAR. The GCN4, Fos, and Jun sequences were chosen to match previously studied peptide models (26, 27). The C/EBP leucine zipper ends at position 340 of rat C/EBP (28), corresponding to the end of a bZIP peptide known to dimerize and bind DNA (29).
GCN4 leucine zippers with different combinations of isoleucines and asparagines were isolated from a pool of mutants constructed by mutually primed DNA synthesis from a pair of oligonucleotides designed to have a random mixture of AAC and ATC codons at four a positions (Fig. 1). From 64 candidates, 40 mutant zippers had changes only at the a positions. Fourteen of the 16 possible sequences were recovered. The remaining 2 sequences were constructed from the recovered sequences by exchanging DNA segments around an internal XhoI site.
Negative Dominance As a Tool to Study Dimerization Specificity.
Specificity assays were done using repressor fusions and dominant-negative mutations by a method similar to methods described by others (30–33) while this work was in progress. Pairwise combinations of fusion proteins with active (cI+) and inactive (cI−) DNA-binding domains were expressed from compatible plasmids in E. coli. Heterodimer formation will result in sensitivity to λ if the level of cI+ homodimers is reduced below the critical level required for phage immunity. To achieve this situation, we expressed cI+ fusions from weak, constitutive promoters, while the cI− fusion proteins were expressed from uninduced Ptac on compatible plasmids. In the experiments described here, the cI− fusions contained the QL44 substitution at the beginning of helix 3, the DNA recognition helix of the helix–turn–helix motif (34). This mutation acts as a dominant-negative allele in intact λ repressor (24).
We set expression of the fusions so that titration occurs whenever the cI+ and dominant-negative fusions contain the same leucine zipper. When immunity is retained, the cI+/cI− heterodimers must be less stable than the cI+ homodimers, since immunity is lost when the leucine zippers are the same. We define each stable homodimer to be specific for itself. For a pair of leucine zippers with different specificities, both retain immunity in the presence of the inhibitor form of the other. Loss of immunity includes inhibition occurring in one direction only and preferential heterodimer formation (35).
RESULTS
Homodimer Formation by a Position Mutants.
Protein fusions to the DNA-binding domain of λ repressor can be used to study the properties of oligomerization domains from other proteins (reviewed in ref. 36). Intact λ repressor, which inhibits growth of phage λ, normally consists of two domains. At low concentrations, the C-terminal domain is required for assembly into homodimers. Removing the C domain renders the protein inactive, while replacing it with a different dimerization domain, such as a leucine zipper, restores dimer formation and repressor activity.
Repressor fusions have been used to characterize the sequence requirements for assembly of the GCN4 leucine zipper (21). Although the immunity phenotype of the repressor fusion system is often insensitive to weakly destabilizing effects of single mutations, these effects can often be observed in combinatorial mutagenesis experiments. For example, combinatorial mutagenesis of the e and g positions in the GCN4 leucine zipper revealed sequence preferences that would not have been observed in single-site mutants (21, 23). Even in the case of the d positions, a strong preference for leucine that was not obvious in single-site mutagenesis was shown by simultaneous, combinatorial mutagenesis of four d positions (23).
To test the tolerance of the a positions to multiple asparagine substitutions, we constructed 16 GCN4 leucine zipper mutants with all combinations of asparagine (N) or isoleucine (I) at the last four a positions. I was used instead of V because codons for I and N differ by only a single change in the second base. Isoleucine has been shown to be stabilizing at the a positions of dimeric coiled coils (18). Each mutant is designated by the amino acids at the mutated positions. For example, NNII has N at positions 9 and 16, and I at positions 23 and 30.
Plasmids were constructed that expressed the mutants at two levels. At the higher level, all but NNNN conferred immunity (Table 1). Thus, leucine zipper mutants with all combinations of 0, 1, 2, or 3 asparagines at the four a positions retain the ability to form oligomers.
Table 1.
Homodimer formation by a position mutants
| No. of Ns | Sequence | Immunity
|
|
|---|---|---|---|
| High expression | Low expression | ||
| 4 | NNNN | s | s |
| 3 | NNNI | i | s |
| NNIN | i | s | |
| NINN | i | i | |
| INNN | i | i | |
| 2 | NNII | i | i |
| NINI | i | i | |
| INNI | i | i | |
| NIIN | i | i | |
| ININ | i | i | |
| IINN | i | i | |
| 1 | NIII | i | i |
| INII | i | i | |
| VNVV | i | i | |
| IINI | i | i | |
| IIIN | i | i | |
| 0 | IIII | i | i |
The number of asparagines and the sequences of the amino acids at positions 9, 16, 23, and 30 are shown on the left. Each mutant was tested at two different expression levels. i Indicates cells that are immune to λ; s indicates sensitivity to λ.
To increase the sensitivity of the assay to disruptive mutations, we decreased the intracellular level of the fusion proteins by changing the promoter in each of the plasmids (see Materials and Methods). At this lower level, 13 of 16 mutants, including all combinations with 0, 1, or 2 asparagines at the four a positions, conferred immunity to phage λ. Three mutants, NNNN, NNIN, and NNNI, were sensitive to λ.
To determine whether the mutant proteins behaved as dimers in vivo, we used reporters that distinguish dimeric fusions from higher-order oligomers (34). In λ112OsPs (37), two λ operators lie upstream of a promoter controlling the cat and lacZ genes. The upstream site has a high affinity for repressor but does not overlap the promoter, while the weaker proximal site represses expression of the reporter genes. λXZ970 has only the weaker site. If a fusion protein forms higher-order oligomers either in solution or after DNA binding, there will be less lacZ expression from λ112OsPs than from λXZ970. By contrast, dimeric fusion proteins will give comparable expression from both reporters. All of the mutants except IIII behaved as dimers in E. coli (data not shown). The IIII mutant repressed lacZ expression from λ112OsPs more than from λXZ970; however, the effect of the upstream operator on repression by IIII was intermediate between our dimeric and tetrameric controls.
Dimerization Specificity Assays.
To study the specificity of heterodimer formation, we developed an assay utilizing dominant-negative (38) repressor fusions (Fig. 2a). Active fusion proteins consist of a leucine zipper fused to the wild-type DNA-binding domain of λ repressor (cI+). For our assay, the cI+ fusion protein must form enough stable homodimers to confer immunity to λ infection when expressed from a constitutive weak promoter in E. coli. An excess of a second fusion protein, consisting of a leucine zipper fused to a mutant form of the DNA-binding domain (cI−), is expressed in the same cell. If the two leucine zippers form heterodimers, the cI− fusion titrates the cI+ fusion into inactive cI−/cI+ heterodimers, reducing the intracellular concentration of functional homodimers, and rendering the cell sensitive to λ. If heterodimers do not form, active homodimers will not be affected by the presence of the cI− fusion protein, and the cell will retain immunity.
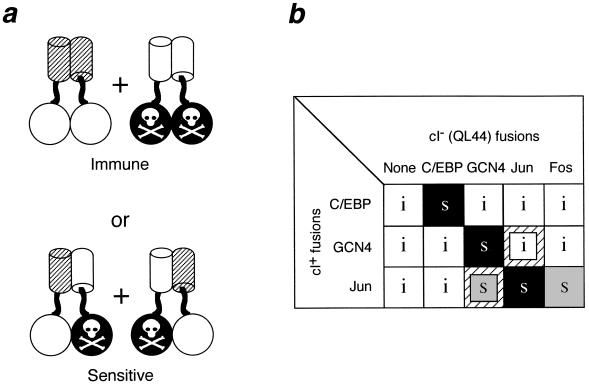
Figure 2.
Rationale for a negative-dominance-based specificity assay with repressor fusions. (a) Active fusions (open circles fused to hatched cylinders) are coexpressed with fusions to an inactivated repressor domain (black circles marked by a skull and crossbones fused to open cylinders). If heterodimers do not form, cells will be immune to phage infection (Upper). Formation of heterodimers will titrate the active monomers into inactive heterodimers, and cells expressing these fusion proteins will be sensitive to killing by phage (Lower). (b) Reconstruction results with the leucine zippers of C/EBP, GCN4, Jun, and Fos. Pairwise combinations of fusion proteins with active (cI+) and inactive (cI−/QL44) DNA-binding domains were expressed from compatible plasmids in E. coli. White boxes marked with an i indicate combinations that are immune to λ; s indicates combinations that are sensitive to λ. Black boxes indicate those pairs where the active and inactive DNA-binding domains are fused to the same leucine zipper; gray boxes indicate titration involving two different leucine zippers. Small inset boxes over a hatched background indicate one-way titrations.
Fig. 2b shows reconstruction experiments with leucine zippers from GCN4, C/EBP, Jun, and Fos. Plasmids expressing fusions to the cI+ repressor domain were introduced into cells in the presence and absence of compatible plasmids expressing fusions to the dominant-negative cIQL44 repressor domain. Since cI+–Fos homodimers are not immune (25), titration was not done using cI+ fusions to the Fos leucine zipper.
cI+–GCN4 is titrated by cIQL44–GCN4, but it retains immunity in the presence of all of the other QL44 fusion proteins. Similarly, C/EBP is titrated only by itself. Jun is titrated by itself and Fos. These results are consistent with the previous description of C/EBP and Fos/Jun as belonging to different specificity classes (39), and they demonstrate that GCN4 and C/EBP are members of noninteracting families. Jun is also titratable by GCN4, while GCN4 is not inhibited by Jun. This asymmetry, which we refer to as “one-way” titration, is consistent with studies showing that the stability of Jun–GCN4 heterodimers, while lower than that of GCN4 homodimers, is comparable to or greater than the stability of Jun homodimers (35, 40).
The reconstruction experiments described above show that the titration assay could detect known differences in the dimerization specificities of leucine zippers. We next used this approach to evaluate the dimerization specificities of the a position mutants. The 13 mutants that conferred immunity at the lower expression level were fused to the inhibitor domain and tested for heterodimerization with mutant and wild-type (VNVV) GCN4. Heterodimers were detected in only 31 of 196 pairwise combinations (Fig. 3). With a few exceptions, the pattern of observed heterodimers is symmetrical around the diagonal formed by the 14 self-interactions, and heterodimers do not form whenever the positions of asparagines differ at the positions 9, 16, and 23. Thus, the majority of the mutants have dimerization specificities different from GCN4 and from each other.
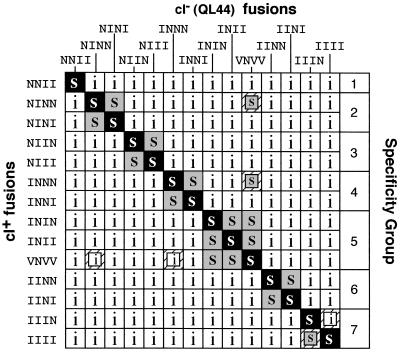
Figure 3.
Effects of a position mutations on dimerization specificity. A subset of the mutants shown in Table 1 were tested for heterodimer formation by the in vivo competition test shown in Fig. 1. Combinations that formed heterodimers and were sensitive to superinfection are indicated by an s. Combinations that retained immunity are indicated by an i. Shadings are as in Fig. 2. Groups of leucine zippers with overlapping specificities are indicated by brackets on the right.
As expected for homodimers, each of the mutant leucine zippers titrates itself. All of the mutants are resistant to titration by most of the other leucine zippers tested. On the basis of the patterns of sensitivity and resistance to titration, the wild-type GCN4 leucine zipper and the 13 mutants can be divided into seven groups such that all of the sequences within a group form heterodimers but are resistant to inhibition by members of the other groups (Fig. 3). In general, heterodimer formation is detected whenever the identities of the amino acids are the same in the first three a positions. Differences at position 30, the a position of the last heptad, do not have a detectable effect on heterodimer formation.
Three pairwise combinations gave results that do not conform to this simple rule (Fig. 3). NINN and INNN are both titratable by GCN4 (VNVV), although formation of heterodimers would require V-N or I-N mismatches to form at several positions. Also, IIIN is resistant to titration by IIII, even though positions 9, 16, and 23 are the same in both mutants.
DISCUSSION
Placement of Buried Asparagines Determines Dimerization Specificity.
Using λ repressor fusion proteins, we examined the dimerization properties of 13 mutants of the GCN4 leucine zipper with different patterns of asparagines and isoleucines at four a positions. These mutants define seven different classes of mutually noninteracting dimers (Fig. 3) and demonstrate that buried asparagines in the dimer interface can determine dimerization specificity. Moreover, the differences in dimerization specificity among the mutants can be understood in terms of a simple physical model.
When a homodimeric leucine zipper assembles as a parallel coiled coil, residues at equivalent a positions are opposed to each other in same layer of the interface (Fig. 1). In the crystal structures of GCN4 leucine zippers (1, 2), the asparagine side chains at position 16 are positioned to form a hydrogen bond across the dimer interface. In NMR studies using a homodimeric Jun leucine zipper peptide, chemical shifts and protection of the γ NH2 protons of Asn-22 against proton exchange suggest that the asparagines at this a position in Jun are in rapid exchange among several hydrogen-bonded conformations (7).
In heterodimers, however, equivalent positions in the sequence may be occupied by different kinds of amino acids. Whenever I or V pairs with N, the N will not be able to form intersubunit side-chain hydrogen bonds that it would form in homodimers. In this case, heterodimers will be less stable than either homodimer.
The most C-terminal a position of the GCN4 leucine zipper (residue 30) is less sensitive to the presence of an unpaired N. This probably reflects fraying at the C termini of the α-helices in the coiled coil. In the crystal structures of the GCN4 leucine zipper, Gly-31 is not in a helical conformation, and Glu-32 and Arg-33 are unstructured (1, 2, 10). Amide protons from Val-30 exchange much faster than those from the other three a positions, and the three residues following Val-30 are largely unprotected from amide proton exchange (41).
Specificity in the pattern of asparagines is also evident in the formation of heterodimers between GCN4 and Jun. In the reconstruction experiment, GCN4 (MVNVV) can titrate Jun (IVNAV) in a one-way fashion. Among the GCN4 a position mutants, only INII and ININ can titrate Jun in a one-way fashion (not shown). This suggests a specific preferred registration of the heptads in the GCN4–Jun heterodimer, with the Asn residues in the third heptads of Jun (IVNAV) and GCN4 (MVNVV), (M)INII, or (M)ININ forming an intersubunit hydrogen bond.
Some of the combinations shown in Fig. 3 cannot be explained by aligning asparagines at the three central a positions. Both NINN and INNN are titrated by GCN4 although the pattern of asparagines would not lead to hydrogen bonding in heterodimers. These mutants do not titrate GCN4, suggesting that the stability of mutant/GCN4 heterodimers is greater than that of the mutant homodimers, but less than that of GCN4 homodimers. Because both of these mutants have asparagines at 3 of 5 a positions, we believe that the NINN and INNN mutants barely form enough homodimers to confer immunity to phage infection. Even a small amount of heterodimer formation with GCN4 would deplete the functional homodimers below the threshold required to give a λ immunity. Note, however, that the NINN and INNN mutants are insensitive to titration by the other a position mutants, including INII, which has the same pattern of polar and nonpolar residues as GCN4 (VNVV). This suggests that V-N mismatches are less destabilizing than I-N mismatches. Alternatively, since the equilibrium among homodimers and heterodimers reflects the relative stabilities of all three species, this difference could reflect differences in the stabilities of the INII and VNVV homodimers.
A different kind of anomaly is seen in the one-way titration of the IIII mutant by the IIIN mutant. IIII fails to titrate IIIN even though they have the same side chains at the first three a positions. Interpreting this result is complicated by the tendency of IIII to form trimers under certain conditions (T. Alber, personal communication) and the possibility that the strongly amphipathic surface of IIII monomers might lead to nonspecific interactions in E. coli.
Effects on Homodimer Stability.
The a and d positions define a 4–3 hydrophobic repeat that forms a nonpolar surface on each of the helices in a leucine zipper. In synthetic peptides, replacing a buried polar group at a position Asn-16 in the GCN4 leucine zipper dramatically increased thermal stability (3, 16), indicating that burying the hydrogen bonded pair of asparagines is destabilizing compared with a pair of β-branched aliphatic residues in an equivalent position in the GCN4 leucine zipper.
In a previous study, it was shown that while the GCN4 leucine zipper tolerates substitutions to other hydrophobic amino acids at individual d positions, the special importance of leucine could be detected in a combinatorial mutagenesis experiment (23). Combinatorial mutagenesis of the four a positions allowing only isoleucine and asparagine revealed a detectable destabilizing effect of inserting asparagines into the dimer interface (Table 1). The magnitude of the effect was surprisingly modest, however, with only the NNNN variant failing to confer immunity at the higher expression level. Among the mutants with three or four asparagines in the a positions, INNN and NINN conferred immunity even at the lower expression level, although on the basis of their ability to repress lacZ reporters (data not shown) and the one-way titration by GCN4, these two homodimers are less stable than the mutants with fewer asparagines.
The destabilizing effects of the asparagines should be compensated to some extent by stabilizing effects of replacing valines with isoleucines (18). Fusion proteins with QNVV and VNVN at the a positions also retained repressor activity at our “high” expression level (23).
Role of the a Positions in Naturally Occurring Leucine Zippers.
Although the pattern of hydrophobic and hydrogen-bonding interactions in the a position is sufficient to define several distinct dimerization specificities, other positions clearly contribute to defining specificity in naturally occurring leucine zippers. Mutants with Leu-to-Val changes at the d positions of Fos and Jun differ in their abilities to form heterodimers (42). Interactions between the surface e and g positions have been shown to be sufficient to explain the specificities of Fos and Jun (35) as well as to design complementary dimers (13). However, the roles of putative interhelical salt bridges formed between charged amino acids in the e and g positions in determining coiled-coil dimerization specificity remain controversial (14, 43–47). We are in the process of applying the assay system described here to characterize how mutations in the e and g positions of GCN4 leucine zipper affect dimerization specificity (X.Z., H. Zhu, H. Lashnel, and J.H., unpublished work).
Does the pattern at the a position contribute to specificity in a biologically significant way? Examining sequences of known leucine zipper proteins suggests that it does. Among 67 leucine zippers (9), asparagine is the most frequent polar amino acid at the a positions and is often found at different heptads. Members of the C/EBP family have asparagine or alanine in the first heptad and asparagine in the third heptad, while the mammalian CREB family has asparagine only in the third heptad. ATF-3 has serine in the second heptad and asparagine in the third. ATF-6 has asparagine in the third and fourth heptads. Our results suggest that sequence differences at the a positions contribute to the ability of these families to form independent regulatory circuits.
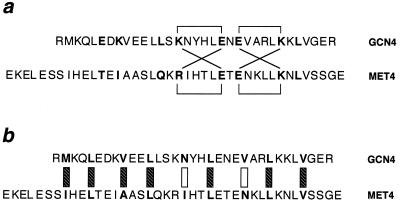
In Saccharomyces cerevisiae, leucine zipper sequences are required for the activity of GCN4 (48) and MET4 (49). These two leucine zippers can be aligned so that charge interactions in the e and g positions in heterodimers and homodimers would be equivalent (Fig. 4). If interactions among the e and g positions were sufficient to allow heterodimers to form in this register, the two proteins might interfere with each other’s activity. However, it is not possible to align the asparagines at the a positions of these two proteins in a plausible heterodimer. Thus the asparagines at the a positions of GCN4 and MET4 may be important for preventing unwanted cross-talk between the two regulatory systems.
Figure 4.
Alignment of the leucine zippers of GCN4 and MET4. (a) Putative salt bridges involving the e and g positions (shown in boldface). Brackets indicate ion pairs observed in GCN4 (above the sequences) and predicted for MET4 (below) homodimers. Diagonal lines indicate similar ion pairs predicted to form in heterodimers. (b) Interactions in the hydrophobic core residues of putative heterodimers. The a and d positions are shown in boldface. Hatched boxes indicate interactions that would not discriminate between homodimers and heterodimers. Open boxes indicate mismatches between hydrophobic and polar side chains.
The pattern of asparagines at the a positions should contribute to identifying permitted and forbidden interactions among other leucine zipper proteins; this will ultimately be important for understanding how these proteins control gene expression in the complex environment of a cell. Other polar residues could influence dimerization specificity in a similar manner. Being able to control the specificity of protein–protein interactions should contribute to efforts toward de novo protein design. Leucine zippers have been used as molecular clamps in engineered proteins (25, 50, 51). Our mutants should be useful for constructing functionally independent, noninteracting systems.
Acknowledgments
We thank Heather Hunter for technical assistance and many colleagues at Texas A&M for helpful suggestions. Tom Alber graciously described unpublished work on the properties of mutants and Carol Gross provided invaluable advice on how to present these data. Marty Scholtz and Debby Siegele provided useful comments on several versions of the manuscript. This work was performed as part of the Ph.D. thesis research of X.Z. and was supported by National Science Foundation Grant MCB-9305403 to J.H.
References
- 1.O’Shea E K, Klemm J D, Kim P S, Alber T. Science. 1991;254:539–544. doi: 10.1126/science.1948029. [DOI] [PubMed] [Google Scholar]
- 2.Ellenberger T E, Brandl C J, Struhl K, Harrison S C. Cell. 1992;71:1223–1237. doi: 10.1016/s0092-8674(05)80070-4. [DOI] [PubMed] [Google Scholar]
- 3.Harbury P B, Zhang T, Kim P S, Alber T. Science. 1993;262:1401–1407. doi: 10.1126/science.8248779. [DOI] [PubMed] [Google Scholar]
- 4.Harbury P B, Kim P S, Alber T. Nature (London) 1994;371:80–83. doi: 10.1038/371080a0. [DOI] [PubMed] [Google Scholar]
- 5.Glover J N M, Harrison S C. Nature (London) 1995;373:257–261. doi: 10.1038/373257a0. [DOI] [PubMed] [Google Scholar]
- 6.Saudek V, Pastore A, Castiglione M M, Frank R, Gausepohl H, Gibson T, Weih F, Roesch P. Protein Eng. 1990;4:3–10. doi: 10.1093/protein/4.1.3. [DOI] [PubMed] [Google Scholar]
- 7.Junius F K, Mackay J P, Bubb W A, Jensen S A, Weiss A S, King G F. Biochemistry. 1995;34:6164–6174. doi: 10.1021/bi00018a020. [DOI] [PubMed] [Google Scholar]
- 8.Hu J, Sauer R. Nucleic Acids Mol Biol. 1992;6:82–101. [Google Scholar]
- 9.Hurst H C. Protein Profile. 1995;2:123–168. [PubMed] [Google Scholar]
- 10.Konig P, Richmond T J. J Mol Biol. 1993;233:139–154. doi: 10.1006/jmbi.1993.1490. [DOI] [PubMed] [Google Scholar]
- 11.Schuermann M, Hunter J B, Hennig G, Müller R. Nucleic Acids Res. 1991;19:739–746. doi: 10.1093/nar/19.4.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O’Shea E K, Lumb K J, Kim P S. Curr Biol. 1993;3:658–667. doi: 10.1016/0960-9822(93)90063-t. [DOI] [PubMed] [Google Scholar]
- 13.Vinson C R, Hai T, Boyd S M. Genes Dev. 1993;7:1047–1058. doi: 10.1101/gad.7.6.1047. [DOI] [PubMed] [Google Scholar]
- 14.Zhou N E, Kay C M, Hodges R S. Protein Eng. 1994;7:1365–1372. doi: 10.1093/protein/7.11.1365. [DOI] [PubMed] [Google Scholar]
- 15.John M, Briand J P, Granger-Schnarr M, Schnarr M. J Biol Chem. 1994;269:16247–16253. [PubMed] [Google Scholar]
- 16.Potekhin S A, Medvedkin V N, Kashparov I A, Venyaminov S Y. Protein Eng. 1994;7:1097–1101. doi: 10.1093/protein/7.9.1097. [DOI] [PubMed] [Google Scholar]
- 17.Lovejoy B, Choe S, Cascio D, McRorie D K, DeGrado W F, Eisenberg D. Science. 1993;259:1288–1293. doi: 10.1126/science.8446897. [DOI] [PubMed] [Google Scholar]
- 18.Zhu B-Y, Zhou N E, Kay C M, Hodges R S. Protein Sci. 1993;2:383–394. doi: 10.1002/pro.5560020310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lumb K J, Kim P S. Biochemistry. 1995;34:8642–8648. doi: 10.1021/bi00027a013. [DOI] [PubMed] [Google Scholar]
- 20.Miller J. Experiments in Molecular Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1972. [Google Scholar]
- 21.Hu J, Newell N, Tidor B, Sauer R. Protein Sci. 1993;2:1072–1084. doi: 10.1002/pro.5560020701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vershon A K, Bowie J U, Karplus T M, Sauer R T. Proteins Struct Funct Genet. 1986;1:302–311. doi: 10.1002/prot.340010404. [DOI] [PubMed] [Google Scholar]
- 23.Hu J C, O’Shea E K, Kim P S, Sauer R T. Science. 1990;250:1400–1403. doi: 10.1126/science.2147779. [DOI] [PubMed] [Google Scholar]
- 24.Nelson H C M, Hecht M H, Sauer R T. Cold Spring Harbor Symp Quant Biol. 1983;47:441–449. doi: 10.1101/sqb.1983.047.01.052. [DOI] [PubMed] [Google Scholar]
- 25.Kim Y-I, Hu J C. Proc Natl Acad Sci USA. 1995;92:7510–7514. doi: 10.1073/pnas.92.16.7510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O’Shea E K, Rutkowski R, Kim P S. Science. 1989;243:538–542. doi: 10.1126/science.2911757. [DOI] [PubMed] [Google Scholar]
- 27.O’Shea E K, Rutkowski R, Stafford W, III, Kim P S. Science. 1989;245:646–648. doi: 10.1126/science.2503872. [DOI] [PubMed] [Google Scholar]
- 28.Landschulz W H, Johnson P F, Adashi E Y, Graves B J, McKnight S L. Genes Dev. 1988;2:786–800. doi: 10.1101/gad.2.7.786. [DOI] [PubMed] [Google Scholar]
- 29.O’Neil K T, Shuman J D, Ampe C, DeGrado W F. Biochemistry. 1991;30:9030–9034. doi: 10.1021/bi00101a017. [DOI] [PubMed] [Google Scholar]
- 30.Bunker C A, Kingston R E. Nucleic Acids Res. 1995;23:269–276. doi: 10.1093/nar/23.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marchetti A, Abril-Marti M, Illi B, Cesareni G, Nasi S. J Mol Biol. 1995;248:541–550. doi: 10.1006/jmbi.1995.0241. [DOI] [PubMed] [Google Scholar]
- 32.Longo F, Marchetti M, Castagnoli L, Battaglia P A, Gigliani F. Biochem Biophys Res Commun. 1995;206:326–334. doi: 10.1006/bbrc.1995.1045. [DOI] [PubMed] [Google Scholar]
- 33.Joung J K, Chung E H, King G, Yu C, Hirsh A S, Hochschild A. Genes Dev. 1995;9:2986–2996. doi: 10.1101/gad.9.23.2986. [DOI] [PubMed] [Google Scholar]
- 34.Zeng X, Hu J C. Gene. 1997;185:245–249. doi: 10.1016/s0378-1119(96)00652-x. [DOI] [PubMed] [Google Scholar]
- 35.O’Shea E K, Rutkowski R, Kim P S. Cell. 1992;68:699–708. doi: 10.1016/0092-8674(92)90145-3. [DOI] [PubMed] [Google Scholar]
- 36.Hu J C. Structure. 1995;3:431–433. doi: 10.1016/s0969-2126(01)00176-9. [DOI] [PubMed] [Google Scholar]
- 37.Beckett D, Burz D S, Ackers G K, Sauer R T. Biochemistry. 1993;32:9073–9079. doi: 10.1021/bi00086a012. [DOI] [PubMed] [Google Scholar]
- 38.Herskowitz I. Nature (London) 1987;329:219–222. doi: 10.1038/329219a0. [DOI] [PubMed] [Google Scholar]
- 39.Lamb P, McKnight S L. Trends Biochem Sci. 1991;16:417–422. doi: 10.1016/0968-0004(91)90167-t. [DOI] [PubMed] [Google Scholar]
- 40.Kouzarides T, Ziff E. Nature (London) 1989;340:568–571. doi: 10.1038/340568a0. [DOI] [PubMed] [Google Scholar]
- 41.Goodman E M, Kim P S. Biochemistry. 1991;30:11615–11620. doi: 10.1021/bi00114a002. [DOI] [PubMed] [Google Scholar]
- 42.Gentz R, Rauscher F D, Abate C, Curran T. Science. 1989;243:1695–1699. doi: 10.1126/science.2494702. [DOI] [PubMed] [Google Scholar]
- 43.Krylov D, Mikhailenko I, Vinson C. EMBO J. 1994;13:2849–2861. doi: 10.1002/j.1460-2075.1994.tb06579.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lumb K J, Kim P S. Science. 1995;268:436–439. doi: 10.1126/science.7716550. [DOI] [PubMed] [Google Scholar]
- 45.Lumb K J, Kim P S. Science. 1996;271:1136. doi: 10.1126/science.271.5252.1137. [DOI] [PubMed] [Google Scholar]
- 46.Lavigne P, Sönnichen F D, Kay C M, Hodges R S. Science. 1996;271:1136. doi: 10.1126/science.271.5252.1136. [DOI] [PubMed] [Google Scholar]
- 47.Zhou N E, Kay C M, Hodges R S. J Mol Biol. 1994;237:500–512. doi: 10.1006/jmbi.1994.1250. [DOI] [PubMed] [Google Scholar]
- 48.Hope I A, Struhl K. Cell. 1986;46:885–894. doi: 10.1016/0092-8674(86)90070-x. [DOI] [PubMed] [Google Scholar]
- 49.Thomas D, Jacquemin I, Surdin K Y. Mol Cell Biol. 1992;12:1719–1727. doi: 10.1128/mcb.12.4.1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Blondel A, Bedouelle H. Protein Eng. 1991;4:457–461. doi: 10.1093/protein/4.4.457. [DOI] [PubMed] [Google Scholar]
- 51.Kostelny S A, Cole M S, Tso J Y. J Immunol. 1992;148:1547–1553. [PubMed] [Google Scholar]