Abstract
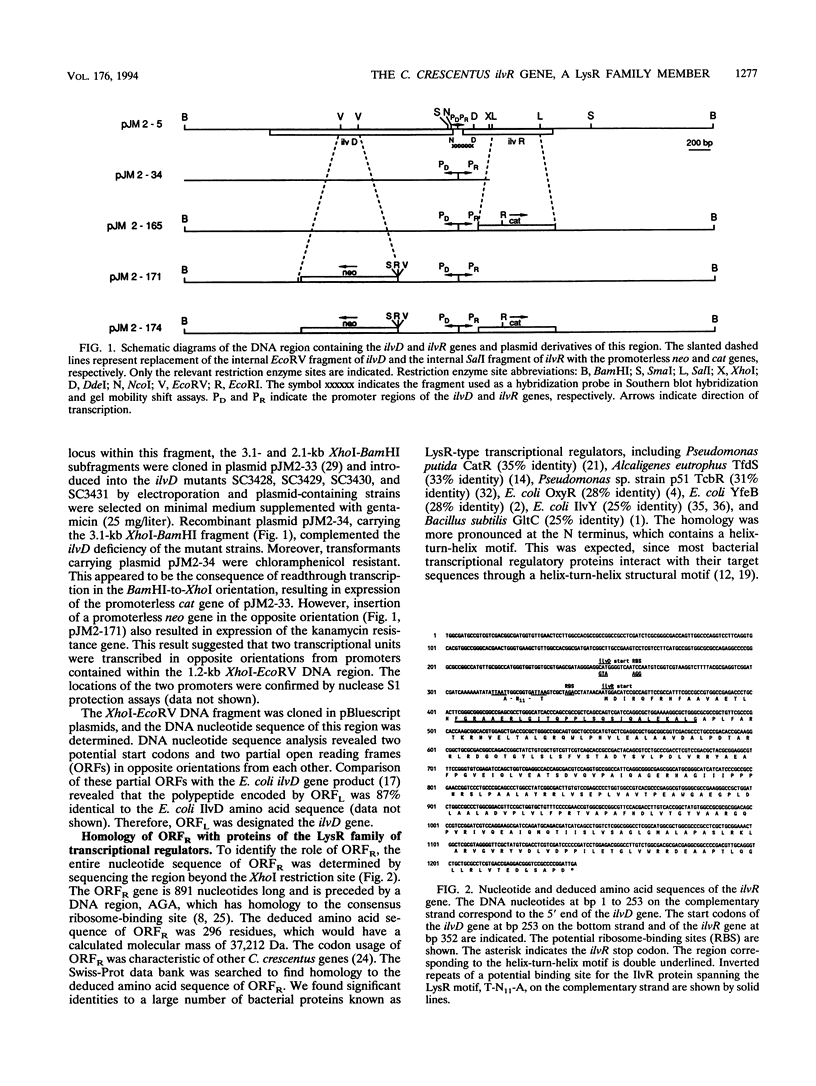
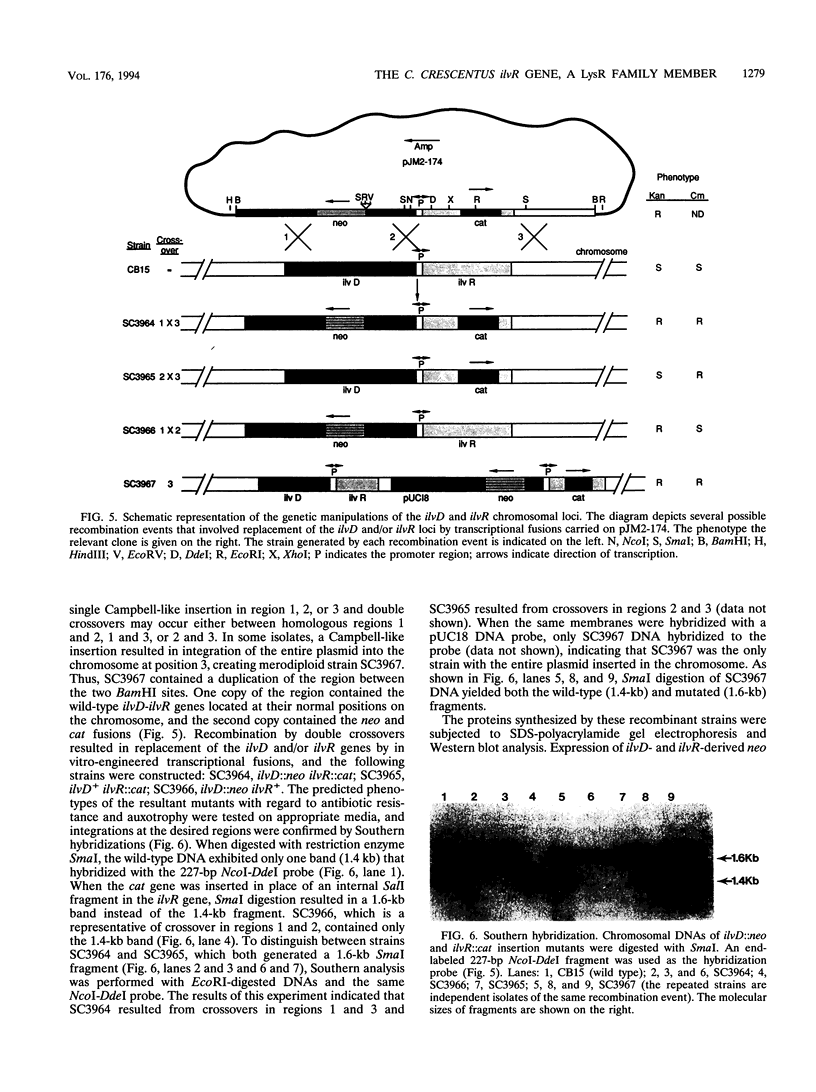
The ilvR gene was located upstream of and transcribed divergently from the ilvD gene of Caulobacter crescentus. DNA nucleotide analysis determined that the ilvR and ilvD translation initiation codons are 98 bp apart. The promoter activity of the DNA region containing the divergent promoters was analyzed by using transcriptional fusions to promoterless reporter genes and immunoblot assays. The results indicate that the ilvR gene product positively regulates the expression of the ilvD gene while negatively autoregulating its own expression. The ilvR gene codes for a protein of 296 amino acid residues (M(r), 37,212). The N-terminal amino acid sequence of the IlvR protein contains a helix-turn-helix motif, suggesting that it is involved in protein-DNA interactions. Protein extracts from both wild-type and merodiploid strains showed specific DNA binding to a 227-bp DNA fragment spanning the ilvD-ilvR promoter region, while no protein-DNA complexes were observed in cell extracts from an ilvR mutant strain. Amino acid sequence comparison revealed that the IlvR protein is a member of the LysR family of transcriptional regulators.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bohannon D. E., Sonenshein A. L. Positive regulation of glutamate biosynthesis in Bacillus subtilis. J Bacteriol. 1989 Sep;171(9):4718–4727. doi: 10.1128/jb.171.9.4718-4727.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brun Y. V., Breton R., Lanouette P., Lapointe J. Precise mapping and comparison of two evolutionarily related regions of the Escherichia coli K-12 chromosome. Evolution of valU and lysT from an ancestral tRNA operon. J Mol Biol. 1990 Aug 20;214(4):825–843. doi: 10.1016/0022-2836(90)90339-N. [DOI] [PubMed] [Google Scholar]
- Chang M., Hadero A., Crawford I. P. Sequence of the Pseudomonas aeruginosa trpI activator gene and relatedness of trpI to other procaryotic regulatory genes. J Bacteriol. 1989 Jan;171(1):172–183. doi: 10.1128/jb.171.1.172-183.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christman M. F., Storz G., Ames B. N. OxyR, a positive regulator of hydrogen peroxide-inducible genes in Escherichia coli and Salmonella typhimurium, is homologous to a family of bacterial regulatory proteins. Proc Natl Acad Sci U S A. 1989 May;86(10):3484–3488. doi: 10.1073/pnas.86.10.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Close T. J., Rodriguez R. L. Construction and characterization of the chloramphenicol-resistance gene cartridge: a new approach to the transcriptional mapping of extrachromosomal elements. Gene. 1982 Dec;20(2):305–316. doi: 10.1016/0378-1119(82)90048-8. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ely B. DNA sequence of the 3' end of the Caulobacter crescentus 16S rRNA gene. Nucleic Acids Res. 1992 Mar 25;20(6):1423–1423. doi: 10.1093/nar/20.6.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ely B. Transfer of drug resistance factors to the dimorphic bacterium Caulobacter crescentus. Genetics. 1979 Mar;91(3):371–380. doi: 10.1093/genetics/91.3.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J., Gussin G. N. Mutations in TrpI binding site II that differentially affect activation of the trpBA promoter of Pseudomonas aeruginosa. EMBO J. 1991 Dec;10(13):4137–4144. doi: 10.1002/j.1460-2075.1991.tb04991.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goethals K., Van Montagu M., Holsters M. Conserved motifs in a divergent nod box of Azorhizobium caulinodans ORS571 reveal a common structure in promoters regulated by LysR-type proteins. Proc Natl Acad Sci U S A. 1992 Mar 1;89(5):1646–1650. doi: 10.1073/pnas.89.5.1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S., Haughn G. W., Calvo J. M., Wallace J. C. A large family of bacterial activator proteins. Proc Natl Acad Sci U S A. 1988 Sep;85(18):6602–6606. doi: 10.1073/pnas.85.18.6602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R. C., Ely B. Isolation of spontaneously derived mutants of Caulobacter crescentus. Genetics. 1977 May;86(1):25–32. doi: 10.1093/genetics/86.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaphammer B., Olsen R. H. Cloning and characterization of tfdS, the repressor-activator gene of tfdB, from the 2,4-dichlorophenoxyacetic acid catabolic plasmid pJP4. J Bacteriol. 1990 Oct;172(10):5856–5862. doi: 10.1128/jb.172.10.5856-5862.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lawther R. P., Lopes J. M., Ortuno M. J., White M. C. Analysis of regulation of the ilvGMEDA operon by using leader-attenuator-galK gene fusions. J Bacteriol. 1990 May;172(5):2320–2327. doi: 10.1128/jb.172.5.2320-2327.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawther R. P., Wek R. C., Lopes J. M., Pereira R., Taillon B. E., Hatfield G. W. The complete nucleotide sequence of the ilvGMEDA operon of Escherichia coli K-12. Nucleic Acids Res. 1987 Mar 11;15(5):2137–2155. doi: 10.1093/nar/15.5.2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neidle E. L., Hartnett C., Ornston L. N. Characterization of Acinetobacter calcoaceticus catM, a repressor gene homologous in sequence to transcriptional activator genes. J Bacteriol. 1989 Oct;171(10):5410–5421. doi: 10.1128/jb.171.10.5410-5421.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- POINDEXTER J. S. BIOLOGICAL PROPERTIES AND CLASSIFICATION OF THE CAULOBACTER GROUP. Bacteriol Rev. 1964 Sep;28:231–295. doi: 10.1128/br.28.3.231-295.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabo C. O., Sauer R. T. Protein-DNA recognition. Annu Rev Biochem. 1984;53:293–321. doi: 10.1146/annurev.bi.53.070184.001453. [DOI] [PubMed] [Google Scholar]
- Rothmel R. K., Aldrich T. L., Houghton J. E., Coco W. M., Ornston L. N., Chakrabarty A. M. Nucleotide sequencing and characterization of Pseudomonas putida catR: a positive regulator of the catBC operon is a member of the LysR family. J Bacteriol. 1990 Feb;172(2):922–931. doi: 10.1128/jb.172.2.922-931.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenlein P. V., Gallman L. S., Winkler M. E., Ely B. Nucleotide sequence of the Caulobacter crescentus flaF and flbT genes and an analysis of codon usage in organisms with G + C-rich genomes. Gene. 1990 Sep 1;93(1):17–25. doi: 10.1016/0378-1119(90)90130-j. [DOI] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. The 3'-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Sung Y. C., Fuchs J. A. The Escherichia coli K-12 cyn operon is positively regulated by a member of the lysR family. J Bacteriol. 1992 Jun;174(11):3645–3650. doi: 10.1128/jb.174.11.3645-3650.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarleton J. C., Ely B. Isolation and characterization of ilvA, ilvBN, and ilvD mutants of Caulobacter crescentus. J Bacteriol. 1991 Feb;173(3):1259–1267. doi: 10.1128/jb.173.3.1259-1267.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbanowski M. L., Stauffer G. V. Genetic and biochemical analysis of the MetR activator-binding site in the metE metR control region of Salmonella typhimurium. J Bacteriol. 1989 Oct;171(10):5620–5629. doi: 10.1128/jb.171.10.5620-5629.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viale A. M., Kobayashi H., Akazawa T., Henikoff S. rbcR [correction of rcbR], a gene coding for a member of the LysR family of transcriptional regulators, is located upstream of the expressed set of ribulose 1,5-bisphosphate carboxylase/oxygenase genes in the photosynthetic bacterium Chromatium vinosum. J Bacteriol. 1991 Aug;173(16):5224–5229. doi: 10.1128/jb.173.16.5224-5229.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Wek R. C., Hatfield G. W. Nucleotide sequence and in vivo expression of the ilvY and ilvC genes in Escherichia coli K12. Transcription from divergent overlapping promoters. J Biol Chem. 1986 Feb 15;261(5):2441–2450. [PubMed] [Google Scholar]
- Wek R. C., Hatfield G. W. Transcriptional activation at adjacent operators in the divergent-overlapping ilvY and ilvC promoters of Escherichia coli. J Mol Biol. 1988 Oct 5;203(3):643–663. doi: 10.1016/0022-2836(88)90199-4. [DOI] [PubMed] [Google Scholar]
- van der Meer J. R., Frijters A. C., Leveau J. H., Eggen R. I., Zehnder A. J., de Vos W. M. Characterization of the Pseudomonas sp. strain P51 gene tcbR, a LysR-type transcriptional activator of the tcbCDEF chlorocatechol oxidative operon, and analysis of the regulatory region. J Bacteriol. 1991 Jun;173(12):3700–3708. doi: 10.1128/jb.173.12.3700-3708.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]