Abstract
We describe a novel method for constructing pools of DNA sequences that encode large proteins with molecular diversity. Sets of primer pairs that form 8 to 10 complementary base pairs in the 3′ region and have double mismatch pairs at their 3′-OH ends were designed so that primer dimers recreated short stretches of DNA (microgenes) devoid of termination codons. Cycles of denaturation and elongation reactions with a pair of primers, four dNTPs, and 3′–5′ exo+ thermostable DNA polymerase gave head-to-tail polymers of the primer dimer unit (microgene) whose sizes exceeded 12 kb. No template was required in this reaction, but mismatched nucleotides at 3′-OH ends of the primers were critical for efficient polymerization. At end-joining junctions of a microgene, nucleotide insertions and deletions randomly occurred, resulting in combinatorial libraries of three reading frames from a single microgene. Further molecular diversity could be incorporated by using a mixture of primers. The resultant polymers have long ORFs whose products have a repetitious nature that could facilitate the formation of higher structures of translated products. Thus, microgene polymers may be used as a source of libraries for in vitro protein evolution experiments. Ligation of a microgene is apparently related to the nonhomologous recombination of double-strand breaks in DNA that has been shown to be catalyzed by DNA polymerases. We named this polymerization reaction the “microgene polymerization reaction.”
Keywords: in vitro evolution, repetitious polypeptides, evolutionary engineering, molecular diversity
A critical step in constructing an in vitro protein evolution system is the generation of a large pool of random DNA sequences that contains long ORFs. Random sequences made from polymerization of four nucleotide blocks (A, T, G, and C) have been used successfully as library sources for many novel functional RNA or DNA molecules (1–4). Such random nucleotide sequences also have been used as a source to generate random amino acid sequences by simply translating them (5). Random DNA sequences, however, contain many triplet codons that serve as translation termination signals (TAA, TAG, and TGA) in their reading frames, which makes the construction of libraries for larger polypeptides more challenging. One possible solution for this problem is to construct “biased” DNA sequences rather than random sequences. For instance, repeats of NNK (where N is A, T, G, or C and K is G or T) can make an ORF that eliminates two out of three termination codons in one reading frame (6, 7). Alternatively, codon-based polymerization can make a single coding frame that is free from termination codons (8–11). These biased DNA sequences, however, still have many termination codons in the remaining coding frames and therefore are very sensitive to frame shifting caused by insertion or deletion. Indeed, 80% of the clones from a codon-based library that was designed to code for a protein of 100 residues has been shown to contain unexpected frame shifting (9).
Our approach to producing libraries with long ORFs is to construct combinatorial polymers from short stretches of DNA (microgenes) rather than using nucleotides as building blocks. The exon theory of genes, proposed by W. Gilbert in 1987 (12), suggested the possibility that new combinations of exons (microgenes) could give birth to novel genes. We have shown that a present day gene can be experimentally dissected into multi-minigene units without loss of its function (13–15), which also suggested that shorter polynucleotide segments could serve as building blocks for generating larger genes. Mimicking the exon shuffling in vitro to create novel proteins has been attempted by in vitro recombination using sexual PCR (16, 17) and by polymerization of microgenes (18–21). These efforts are expected to establish a new system for in vitro protein creation.
In this report, we introduce another type of microgene-based library construction. This approach uses only one microgene as a building block. A microgene is polymerized in a head-to-tail manner using the technique called “microgene polymerization reaction” (MPR) that we established in this study. Nucleotide insertion or deletion randomly occurs at end-joining junctions in MPR, which changes the reading frame of the microgene at junctions. Thus, the resultant microgene polymers are combinatorial libraries of 2 × 3 reading frames of a microgene. When a starting microgene does not contain termination codons in its six frames, the resultant polymers are also free of termination codons, permitting the construction of DNA libraries containing large ORFs.
MATERIALS AND METHODS
MPR.
A 50-μl reaction mixture contained 20 pmol of each of the primers, 350 μM of dNTPs (Amersham), 50 mM Tris·HCl (pH 9.2), 1.75 mM MgCl2, 14 mM (NH4)2SO4, and 2.6 units of Expand Taq polymerase mixture (Boehringer Mannheim). Vent (wt; 2.6 units) or Vent (exo−) DNA polymerase (New England Biolabs) also was used in 20 mM Tris·HCl (pH 8.8), 2 mM MgSO4, 10 mM (NH4)2SO4, 10 mM KCl, and 0.1% Triton X-100. Standard cycling conditions for a System 2400 or 9600 PCR cycler (Perkin–Elmer) were 94°C for 10 min, 63–69°C for 10 min, 35–65 cycles of 94°C for 10 sec, and 63–69°C for 1 min. Enzyme was added after the temperature reached at 94°C. The last incubation was extended to 7 min.
Analyses of Translated Products from Microgene–Polymers.
Amplified DNAs were cloned into the SmaI site of pKS575, which is a derivative of pET19b (22) (Novagen), and the SmaI cloning site and universal translation terminator sequences (TTAATTAATTAA) were inserted after an oligo histidine–tag sequence (23) (Y.T., unpublished work). One of six coding frames of a cloned microgene–polymer was transcribed from a T7 promoter on the plasmid and was translated as a fusion protein with a histidine–tag polypeptide. Ten clones were chosen randomly from each polymer, and their proteins were induced with 0.1 mM isopropyl β-d-thiogalactopyranoside for 3 h at 37°C in strain BL21(DE3) (22). Total cell extracts were analyzed by SDS/PAGE on a 10–20% gradient gel (Dai-ichi Pure Chemicals, Tokyo). Molecular mass markers were run in one lane (Dai-ichi Pure Chemicals).
RESULTS
MPR.
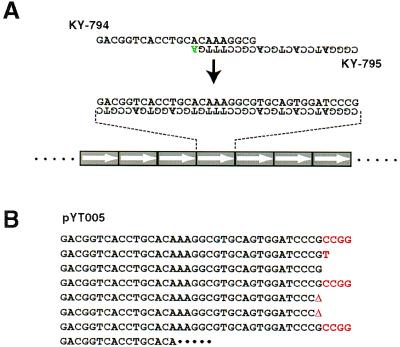
The polymerization of a short stretch of DNA (microgene) was first observed when we performed PCR with a primer set of KY-794 and KY-795 (Fig. 1A) under long PCR conditions (24). These primers form eight complementary base pairs in the 3′ region and have a single extra nucleotide at the 3′-OH end (of KY-795) that does not pair with KY-794 (Fig. 1A). These primers gave a heterogeneous “smeared” DNA product without any template DNA after 35 cycles of PCR with all four dNTPs and an enzyme mixture of Taq and Pwo polymerases (a similar result is shown in Fig. 2B). The sizes of these heterogeneous DNA products ranged from ≈500 bp to >12 kb, and increased cycle numbers resulted in the production of a larger population of DNA (data not shown). We were interested in the origin of these heterogeneous DNA molecules and determined the nucleotide sequences after cloning them into a plasmid vector. The cloned products were shown to comprise tandem repeats of a short stretch of DNA (Fig. 1A). This 36-bp repeating unit is a primer dimer product (25) of KY-794 and KY-795.
Figure 1.
Head-to-tail tandem repeats formation in MPR. (A) Primers used in MPR, primer dimer, and schematic representation of head-to-tail tandem polymer of primer dimer. Mismatched nucleotide at 3′-OH end is in green. (B) An example of a microgene polymer (pYT005). Inserted nucleotides or deletion (Δ) at junction points of a microgene unit is in red.
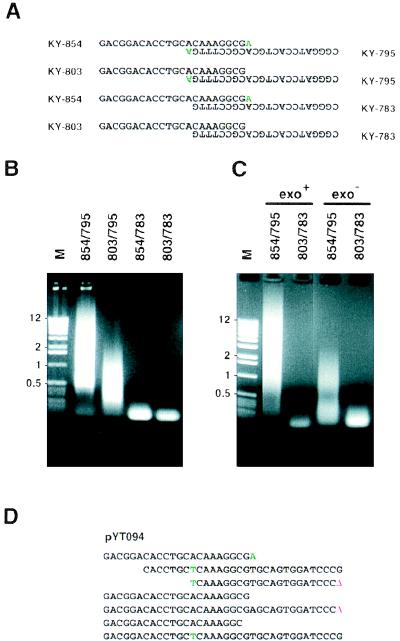
Figure 2.
Effects of the 3′-OH mismatch and 3′–5′ exonuclease activity of DNA polymerase on MPR. (A) Four sets of primers that have double, single, single, and no 3′-OH mismatch nucleotide(s). Mismatch nucleotides are in green. (B) MPR product (10 μl) was fractionated through a 1.2% agarose gel. Lane M, DNA size marker (1-Kb DNA ladder; Life Technologies, Gaithersburg, MD). The sizes of some fragments are indicated on the left (in kilobases). Forty-five cycles of MPR were performed, and the annealing temperature was 69°C. (C) Vent (wt) or Vent (exo−) DNA polymerase was used instead of the Expand Taq polymerase mixture, 45 cycles of MPR were performed, and the annealing temperature was 69°C. (D) An example of a microgene polymer (pYT094) created from KY-854 and KY-795 with Vent (exo−) DNA polymerase. Deletions (Δ) at junctions are in red. Nucleotides originating from the 3′-OH mismatches are in green.
The repeating unit polymerized in a head-to-tail manner without exception as far as examined and was accompanied by insertion or deletion of a few nucleotides at their junctions. In one example (Fig. 1B) among seven junctions, four had insertions of one or four nucleotides, two had a single nucleotide deletion, and one junction had no insertion/deletion. Although a tetra-nucleotide CCGG was inserted frequently at junctions from many clones, other inserted nucleotides included a single T or C or the trinucleotides CGG, GCG, and CCA (data not shown). These insertion sequences may not have originated from the primers because the tetranucleotide CCGG (or its complement GGCC) is not contained in the sequences of these primers.
Primers KY-794 and KY-795 were prepared on a DNA synthesizer and were purified by HPLC. To exclude the possibility that the observed polymer formation was due to unexpected modification of the primers during DNA synthesis or HPLC purification, another set of primers with the same sequences as KY-794 and KY-795 was synthesized using another DNA synthesizer and was purified on a NAP-10 gel filtration column (Pharmacia Biotech, Uppsala, Sweden). These newly synthesized primers gave similar polymers of a primer dimer (data not shown), suggesting that the polymerization was not due to aberrant synthesis or unexpected modification of KY-794 and KY-795 primers. Polymers of a primer dimer were obtained reproducibly with these primers under the conditions described, and we named this reaction the microgene polymerization reaction (MPR).
Factors That Affect Polymerization Efficiency in MPR.
To determine factors that gave efficient polymerization of primer dimers in MPR with KY-794 and KY-795, we performed the following experiments. First, we focused on the single extra nucleotide at the 3′-OH end of KY-795 that does not pair with KY-794 (Fig. 1A). To assess the contribution of the mismatch at 3′-OH ends of primers on MPR, we prepared four pairs of primers that have double, single, and no mismatch pair(s) at their 3′-OH termini (Fig. 2A). After 45 cycles of MPR with a mixture of Taq and Pwo polymerases, the primer pair with double mismatches (KY-854/795) produced the most diffuse smear of DNA whereas primers that lacked mismatches at their 3′-OH ends (KY-803/783) did not produce smeared DNA in these conditions (Fig. 2B). These results indicated that mismatched base pairs at primer 3′-OH termini are a necessary factor in MPR. The effects of a single mismatch were dependent on the nucleotide sequence of the primer. A single base mismatch at the 3′-OH terminus of KY-795 led to the production of smeared DNA whereas the mismatch on KY-854 did not produce a polymer under similar conditions. The factors that caused this difference are not known.
Second, we tested the effect of different polymerases on MPR. The long PCR protocol used in the initial experiments contained a mixture of thermostable Taq and Pwo DNA polymerases. The former enzyme has 5′–3′ exonuclease activity in its N-terminal domain (26) but lacks detectable 3′–5′ exonuclease activity (27) whereas the latter, archaeal DNA polymerase has 3′–5′ exonuclease activity but is believed to lack 5′–3′ exonuclease activity (28). Because no heterogeneous “smeared” DNA was observed with the primers when Taq polymerase was used in standard PCR conditions, we reasoned that the 3′–5′ exonuclease activity of the Pwo DNA polymerase was critical to MPR. Several 3′ exonuclease plus polymerases and their exonuclease minus derivatives are commercially available. Vent DNA polymerase originates from the hyperthermophilic archaeal bacterium Thermococcus litoralis, and its 3′ exonuclease deficient mutant, Vent exo−, has been constructed (28). We next performed MPRs using these Vent wt and Vent exo− polymerases instead of a mixture of Taq and Pwo polymerases with primer pairs KY-854/795 (double mismatches) or KY-803/783 (no mismatch). The MPR with these polymerases gave similar results as the MPR with a mixture of Taq and Pwo polymerases (Fig. 2C). Primers without mismatch pairs at the 3′-OH ends (KY-803/783) did not produce polymers in the presence or absence of 3′–5′ exonuclease activity, confirming the importance of the 3′-OH end mismatch. The combination of primers with double mismatches (KY-854/795) and Vent (exo+) DNA polymerase resulted in the abundant production of heterogeneous DNA, indicating that a Taq polymerase that has 5′–3′ exonuclease activity is not necessarily required for polymer formation. An unexpected result was the production of polymers from the combination of primers with double mismatches (KY-854/795) and Vent (exo−) DNA polymerase although the amount of products was much less compared with that with Vent wt polymerase. Because primer dimers are formed by elongation from one primer using the other primer as template (25), the mismatched 3′ ends of KY-854/795 should be removed by the 3′–5′ exonuclease activity before chain elongation. We were interested in the structure of heterologous DNA products from MPR with KY-854/795 and Vent (exo−) and so determined the nucleotide sequences of them. The polymers were made essentially from sequences derived from two primers (KY-854/795), but additional sequence irregularities were present (one example is shown in Fig. 2D). In these cases, primer dimers were not necessarily repeating units, but, instead, various shorter sequences that were related to the primer dimer were randomly joined. In some cases, nucleotide sequences that originated from the 3′ ends of two primers were observed in the sequence, indicating that the aberrant chain extension had occurred in MRP. In some joining junctions, one primer seemed to connect directly to itself or a primer dimer.
From the above results, we concluded that the factors that enhance the efficiency of MPR are (i) the existence of mismatched nucleotides at 3′-OH ends of primers and (ii) 3′–5′ exonuclease activity of DNA polymerase. The role of the mismatched nucleotide at 3′-OH ends on MPR is further discussed below.
MPR with Mixed Primers Increases Molecular Diversity of Polymers.
The aim of our study was to construct a pool of protein-coding DNAs with a high degree of molecular diversity that can serve as a starting point for in vitro protein evolution experiments. Polymerization of microgenes that are deficient in termination codons is one approach to constructing such a library. MPR can be used for this purpose if the primers are designed so that the primer dimers correspond to a stop-codon-free repeating unit. Randomly inserted nucleotides or deletions at junctions alter the reading frame of a microgene in every repeating unit, and the resultant MPR products are a combinatorial library made from 2 × 3 reading frames. In addition to this molecular diversity resulting from frame switching, we tested the possibility that diversity could be further acquired by using a mixture of primers in MPR.
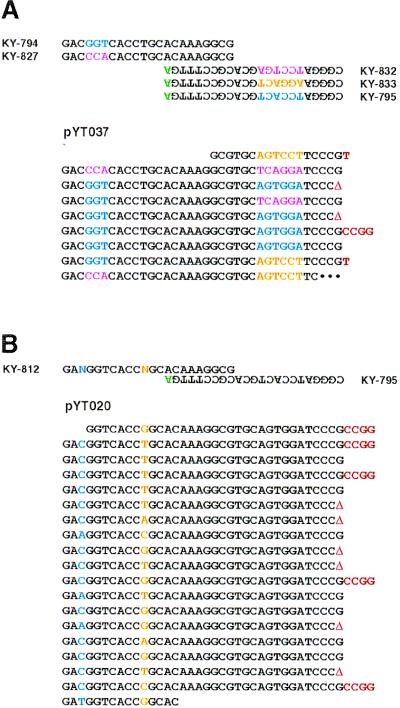
First, we used five primers in MPR instead of two. These included the initial KY-794 and KY-795 and their three derivatives, whose sequences differ from their parent primer at three positions (Fig. 3A). Sequence analyses of cloned polymers from MPR with these primers revealed that polymers were made from mixtures of the five primers (one example is shown in Fig. 3A). Similarly, a primer that was degenerate at specific positions was used to increase molecular diversity. KY-812, a derivative of KY-794, has two degenerate positions (Fig. 3B). MPR with KY-812 and KY-795 gave polymers with two positions randomized (one example is shown in Fig. 3B). It should be noted that the first degenerate position (the third nucleotide of KY-794) had a preference for C (Fig. 3B) whereas the second degenerate position (the 11th nucleotide of KY-794) did not have a preference in this experiment. Thus, usage of mixed primers or degenerated primers in MPR increases the molecular diversity of the resultant polymers.
Figure 3.
MPR with mixtures of primers. (A) Five primers used in MPR are shown on the top. Mismatched nucleotides are in green, and sequences that are differ among primers are separately colored. MPR was carried out with an Expand Taq polymerase mixture for 45 cycles at 69°C for extension. An example of a microgene polymer (pYT037) is shown on the bottom. Inserted nucleotides or deletion (Δ) at junctions is in red. (B) A degenerated primer was used in MPR. Ns represent a mixture of A, T, G, and C. MPR was carried out with an Expand Taq polymerase mixture for 65 cycles at 69°C for extension. An example of a polymers (pYT020) is shown.
Designer Microgenes.
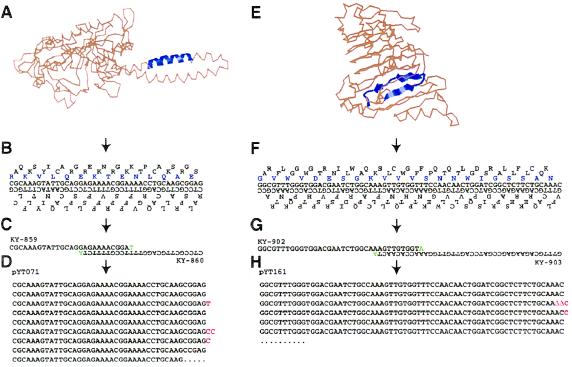
Although a high degree of molecular diversity can be generated from random frame switching at junction points and the use of primer mixtures in MPR, we expected that the nature of the resultant libraries would be greatly influenced by the choice of the initial microgene. To show that the MPR technique is applicable to a wide variety of microgenes, we designed two microgenes starting from polypeptide sequences found in proteins with known three-dimensional structure and constructed libraries through MPR. One microgene had 42 bp and was designed from a 14-amino acid peptide that constituted a part of a coiled coil α-helix in seryl-tRNA synthetase (29) (Fig. 4A). The microgene was designed so that one frame codes for the 14-amino acid polypeptide and the other five frames do not contain any termination codons. The second 66-bp microgene was designed from one turn of a parallel β-helix protein, pectate lyase from Erwinia chrysanthemi (32). For this microgene, a 22-amino acid consensus sequence was first designed from the repeating units, each of which is comprised of three β-strands (Fig. 4B). Primer pairs were designed for these two microgenes so that (i) primer pairs could form 10 and 9 base pairs in their 3′ regions; (ii) the sequences of primer dimers could recreate microgene sequences; and (iii) mismatched single extra nucleotides (A or T) were added at their 3′-OH ends. MPR with these primers and Vent wt DNA polymerase produced heterologous, smeared DNA as expected, and sequence analyses of cloned products revealed that the microgenes were tandemly polymerized with nucleotide insertions or deletions at junctions (Fig. 4 D and H).
Figure 4.
Examples of designer microgenes. (A) Sequence of 14 amino acids was extracted from the anti-paralleled coiled coil α-helix region of the N-terminal domain of Escherichia coli seryl-tRNA synthetase (30). The structure of the enzyme from T. thermophilus is shown (29), and the corresponding region in this enzyme is represented in blue [drawn using Rasmol (31)]. (B) From the extracted sequence, a microgene was designed so that one of six frames codes for the extracted sequence and the others do not contain any termination codons. The sixth residue (Val) in the extracted sequence had to be changed to Glu in the microgene to avoid the appearance of a termination codon in other frames. (C) MPR primers were designed for the microgene. Mismatched nucleotides were added at 3′-OH ends of both primers (in green). (D) An example of a microgene polymer (pYT071) made from KY-859 and KY-860. MPR was carried out for 65 cycles using Vent (exo+) DNA polymerase. The annealing and extension temperature was 63°C. Inserted nucleotides at junctions are in red. (E–H) Design of a microgene from a parallel β-helix protein, E. chrysanthemi pectate lyase E (32), and an example of a microgene polymer. The strategy was same as for A–D, except the 22-amino acid sequence was a consensus sequence comprised of three β-strands.
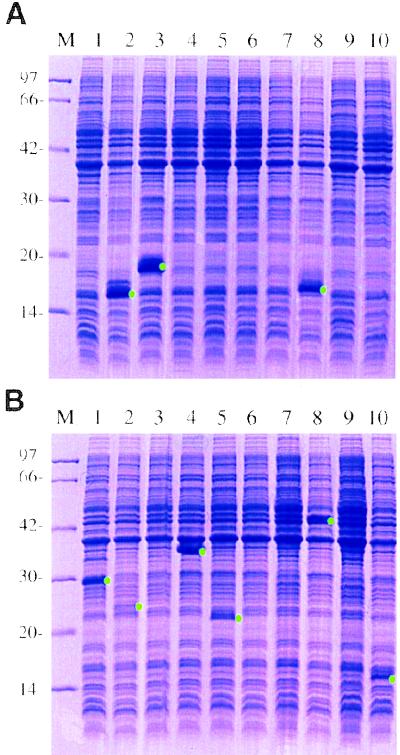
Polymers made from designed microgenes were cloned into an expression vector, and one reading frame was translated in E. coli. Expressed proteins were analyzed by SDS/PAGE. Examples of 10 clones for each microgene are shown (Fig. 5). Three clones from the α-helix microgene and six clones from the β-strand microgene, whose sizes of polymer inserts ranged from 0.25 to 0.8 kb, produced polypeptides with apparent molecular masses of 15–44 kDa, indicating that the MPR products can serve as a source for in vitro protein evolution experiments. We are now analyzing physical characters of the polypeptides translated from microgene polymers.
Figure 5.
SDS/PAGE of proteins from E. coli cells expressing one reading frame of microgenes polymers. (A) α-Helix microgene. (B) β-Strand microgene. Molecular markers were run in lane M, and their sizes are shown on the left in kilodaltons. Visible bands that are expected to be products are indicated by green dots. Some of them (clones 3 and 8 from A and 4, 5, and 8 from B) were confirmed to be products of microgene by purifying them using oligo histidine–tag (data not shown).
DISCUSSION
The repetitious nature of coding sequences has been pointed out by Ohno (33), and from these observations, he has proposed that genes have arisen from repeats of short nucleotide sequences (33–35). Oligomeric repeats have the following advantages as a source of protein-encoding sequences. First, they could contain rather large ORFs and could be relatively tolerant to frame shifting caused by deletions and insertions, if a repeating sequence is devoid of termination codons (34). The probability of generating large protein-encoding genes from random sequences made from A, T, G, and C is theoretically very low because of the appearances of termination codons (34). Second, proteins translated from repeated DNA sequences have periodical amino acid sequences and consequently might be predicted to have higher propensities to form secondary structures (34). Indeed, de novo-designed repetitive polypeptides, including ((GlyAla)3GlyGlu)n, have been shown to form stable secondary structures (36, 37), and such repetitive polypeptides have been receiving attentions in material science (38, 39).
The MPR strategy described here is a simple method to create tandem repeats of microgenes. Fire and Xu (40) have reported an elegant method to make repeated sequences by mimicking rolling circle replication. Their method, however, requires multiple steps to constructing polymers, including phosphorylation, ligation, rolling circle replication, and synthesis of complementary strand. In contrast, MPR requires a rather simple protocol. Furthermore, the resultant polymers are combinatorial libraries made from three reading frames because random insertion and deletion occur at junctions. For example, a 120-amino acid protein library (360 bp) made from a 36-bp microgene could have 2 × 310 = ≈1.2 × 105 molecular diversity. If we take into account deletion and insertion at junction points, the diversity of the population is much greater. Also, additional diversity can be incorporated by using a mixture of primers in MPR, as shown above (Fig. 3).
The choice of the starting microgene will influence the nature of the resultant library. We presently do not have sufficient data to predict the features of a microgene that will translate into an optimal library. Here, we show one example of a design strategy, that is, designing a microgene from a secondary structural element of an existing protein. The rationale of this approach is that at least one–sixth of the sequences in the microgene should code for polypeptides that could form secondary structures; this enhances the possibility that the library contains genes for folded proteins. Different types of folding pattern could emerge from polymerization of secondary structure elements, and this could not be expected from polymerization of individual domains. We designed two microgenes that are expected to code for α-helix-forming and β-strand-forming sequences. Six clones out of 10 randomly selected polymers from the β-strand microgene produced a large amount of proteins, and three out of four proteins investigated have been shown to be soluble (data not shown), suggesting that the polymers have ordered structures. It also should be noted that apparent molecular masses of some clones were higher than ones expected from the DNA sequences (for instance, clone 5 in Fig. 5 produced a polypeptide with an apparent molecular mass of 23 kDa, but its calculated value was 12.7 KDa). Similar anomalous migration on SDS/PAGE also was observed in a β-sheet-forming, repeated polypeptide (36). Details of biochemical characterization will be reported elsewhere. We also are designing microgenes by other approaches, including designing from repeated nucleotide sequences of existing genes and designing genes whose products from all six frames are predicted theoretically to form secondary structural elements.
Although the understanding of the underlying mechanism of MPR awaits further studies, the reaction apparently is related to illegitimate recombination (nonhomologous recombination) in which double-strand breaks of DNA are joined. It has been shown that joining between (i) blunt end DNA and a 3′ protruding single strand (PSS), (ii) blunt end DNA and 5′ PSS, and (iii) 5′ PSS and 3′ PSS can be achieved by a Xenopus egg extract that lacked single-strand-specific DNA ligase activity (41). From these results, Thode et al. (41) postulated the existence of “an alignment protein” that juxtaposes two DNA ends so that DNA synthesis can proceed on a discontinuously aligned DNA (41). DNA polymerases such as the Klenow fragment of DNA polymerase I and Taq polymerase have been shown to serve as the alignment protein themselves (42–44). In vitro systems that contained only a DNA polymerase, four dNTPs, and substrate DNAs were able to synthesize polynucleotides that were replicated from two discontinuous template DNAs (42–44). A critical event in MPR to expand repeats of primer dimers must be the formation of a head-to-tail dimer of a primer dimer because once such tandem repeats are formed, the repeats can expand rapidly in replication cycles (35). We believe that the formation of an initial “dimer of a primer dimer” in MPR is catalyzed by the end-joining activity of a DNA polymerase. Nucleotide insertions may be the result of the joining reaction between oligonucleotides and dNTP monomers (45, 46). The enzyme should make both head-to-tail and head-to-head dimers of primer dimer in the initial ligation, but the former dimer may overgrow in the following cycles.
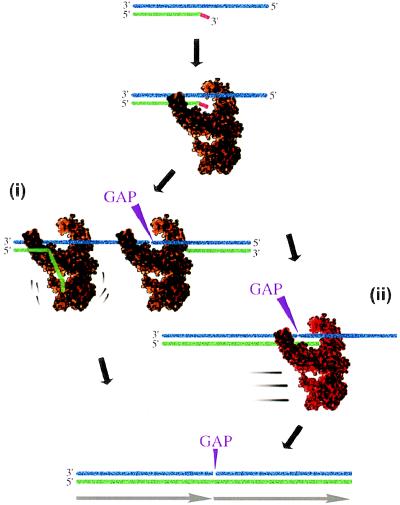
Our results showed that the 3′-OH mismatched nucleotides were important for efficient polymerization. We propose two models for the role of these mismatched nucleotides. In the first model, formation of an editing complex (47) consisting of a template DNA, a primer DNA, and a DNA polymerase delays the synthesis of duplex DNA and increases the probability that the polymerase will juxtapose a 5′-OH end of one template and a 3′-OH of another template (Fig. 6 Left). In the second model, a putative repairing mode of DNA polymerase is postulated. This enzyme would have an increased activity for joining double-strand breaks and would be induced by a 3′-OH mismatched nucleotide (Fig. 6 Right).
Figure 6.
Possible mechanisms in which mismatched nucleotides at the 3′-OH end contribute to the formation of a dimer of primer dimer. Template DNA and primer DNA are shown in blue and green, respectively. A 3′-OH mismatched nucleotide on a primer is in red. In model (i), formation of an editing complex consisting of a template, a primer, and DNA polymerase (shown in brown) increases the likehood that a polymerase will juxtapose a 5′-OH end of one template and a 3′-OH end of another template. In model (ii), a putative repair mode of enzyme (shown in light brown) is postulated that could catalyze both editing and nonhomologous recombination more efficiently than the standard mode of polymerase action. In both cases, DNA polymerase synthesizes repeated units on templates that have phosphate breakage (indicated by GAP) to result in end-joining of a primer dimer unit.
Acknowledgments
We thank T. Hatada for her involvement in the initial phase of this work and W. T. Miller for his critical reading of the manuscript.
ABBREVIATION
- MPR
microgene polymerization reaction
References
- 1.Ellington A D, Szostak J W. Nature (London) 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 2.Tuerk C, Gold L. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 3.Bock L C, Griffin L C, Latham J A, Vermaas E H, Toole J J. Nature (London) 1992;355:564–566. doi: 10.1038/355564a0. [DOI] [PubMed] [Google Scholar]
- 4.Bartel D P, Szostak J W. Science. 1993;261:1411–1418. doi: 10.1126/science.7690155. [DOI] [PubMed] [Google Scholar]
- 5.Dube D K, Loeb L A. Biochemistry. 1989;28:5703–5707. doi: 10.1021/bi00440a001. [DOI] [PubMed] [Google Scholar]
- 6.Cwirla S E, Peters E A, Barrett R W, Dower W J. Proc Natl Acad Sci USA. 1990;87:6378–6382. doi: 10.1073/pnas.87.16.6378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scott J K, Smith G P. Science. 1990;249:386–390. doi: 10.1126/science.1696028. [DOI] [PubMed] [Google Scholar]
- 8.Glaser S M, Yelton D E, Huse W D. J Immunol. 1992;149:3903–13. [PubMed] [Google Scholar]
- 9.Davidson A R, Sauer R T. Proc Natl Acad Sci USA. 1994;91:2146–2150. doi: 10.1073/pnas.91.6.2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davidson A R, Lunb K J, Sauer R T. Nat Struct Biol. 1995;2:856–864. doi: 10.1038/nsb1095-856. [DOI] [PubMed] [Google Scholar]
- 11.Auld D S, Schimmel P. Science. 1995;267:1994–1996. doi: 10.1126/science.7701322. [DOI] [PubMed] [Google Scholar]
- 12.Gilbert W. Cold Spring Harbor Symp Quant Biol. 1987;52:901–905. doi: 10.1101/sqb.1987.052.01.098. [DOI] [PubMed] [Google Scholar]
- 13.Shiba K, Schimmel P. Proc Natl Acad Sci USA. 1992;89:1880–1884. doi: 10.1073/pnas.89.5.1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shiba K, Schimmel P. J Biol Chem. 1992;267:22703–22706. [PubMed] [Google Scholar]
- 15.Shiba K. In: Tracing Biological Evolution in Protein and Gene Structures. Go M, Schimmel P, editors. Amsterdam: Elsevier; 1995. pp. 11–21. [Google Scholar]
- 16.Stemmer W P C. Nature (London) 1994;370:389–391. doi: 10.1038/370389a0. [DOI] [PubMed] [Google Scholar]
- 17.Stemmer W P C. Proc Natl Acad Sci USA. 1994;91:10747–10751. doi: 10.1073/pnas.91.22.10747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nord K, Nilsson J, Nilsson B, Uhlen M, Nygren P A. Protein Eng. 1995;8:601–608. doi: 10.1093/protein/8.6.601. [DOI] [PubMed] [Google Scholar]
- 19.Shiba K, Hatada T, Noda T. Protein Eng. 1996;9:813–814. [Google Scholar]
- 20.Fisch I, Kontermann R E, Finnern R, Hartley O, Solergonzalez A S, Griffiths A D, Winter G. Proc Natl Acad Sci USA. 1996;93:7761–7766. doi: 10.1073/pnas.93.15.7761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mikheeva S, Jarrell K A. Proc Natl Acad Sci USA. 1996;93:7486–7490. doi: 10.1073/pnas.93.15.7486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Studier F W, Rosenberg A H, Dunn J J. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 23.Van Dyke M W, Sirito M, Sawadogo M. Gene. 1992;111:99–104. doi: 10.1016/0378-1119(92)90608-r. [DOI] [PubMed] [Google Scholar]
- 24.Barnes W M. Proc Natl Acad Sci USA. 1994;91:2216–2220. doi: 10.1073/pnas.91.6.2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gelfand D H, White T J. In: PCR Protocols: A Guide to Methods and Applications. Innis M A, Gelfand D H, Sninsky J J, White T J, editors. San Diego: Academic; 1990. pp. 129–141. [Google Scholar]
- 26.Barnes W M. Gene. 1992;112:29–35. doi: 10.1016/0378-1119(92)90299-5. [DOI] [PubMed] [Google Scholar]
- 27.Tindall K R, Kunkel T A. Biochemistry. 1988;27:6008–6013. doi: 10.1021/bi00416a027. [DOI] [PubMed] [Google Scholar]
- 28.Kong H, Kucera R B, Jack W E. J Biol Chem. 1993;268:1965–1975. [PubMed] [Google Scholar]
- 29.Cusack S, Berthet-Colominas C, Härtlein M, Nassar N, Leberman R. Nature (London) 1990;347:249–255. doi: 10.1038/347249a0. [DOI] [PubMed] [Google Scholar]
- 30.Hartlein M, Madern D, Leberman R. Nucleic Acids Res. 1987;15:1005–1017. doi: 10.1093/nar/15.3.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sayle R A, Milner-White E J. Trends Biochem Sci. 1995;20:374–376. doi: 10.1016/s0968-0004(00)89080-5. [DOI] [PubMed] [Google Scholar]
- 32.Yoder M D, Lietzke S E, Jurnak F. Structure (London) 1993;1:241–251. doi: 10.1016/0969-2126(93)90013-7. [DOI] [PubMed] [Google Scholar]
- 33.Ohno S. Proc Natl Acad Sci USA. 1981;78:7657–7661. doi: 10.1073/pnas.78.12.7657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ohno S, Epplen J T. Proc Natl Acad Sci USA. 1983;80:3391–3395. doi: 10.1073/pnas.80.11.3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ohno S. J Mol Evol. 1987;25:325–329. doi: 10.1007/BF02603117. [DOI] [PubMed] [Google Scholar]
- 36.Krejchi M T, Atkins E D T, Waddon A J, Fournier M J, Mason T L, Tirrell D A. Science. 1994;265:1427–1432. doi: 10.1126/science.8073284. [DOI] [PubMed] [Google Scholar]
- 37.McGrath K P, Fournier M J, Mason T L, Tirrell D A. J Am Chem Soc. 1992;114:727–733. [Google Scholar]
- 38.Tirrell D A, Fournier M J, Mason T L. Curr Opin Struct Biol. 1991;1:638–641. [Google Scholar]
- 39.Ball P. Nature (London) 1994;367:323–324. doi: 10.1038/367323a0. [DOI] [PubMed] [Google Scholar]
- 40.Fire A, Xu S-Q. Proc Natl Acad Sci USA. 1995;92:4641–4645. doi: 10.1073/pnas.92.10.4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thode S, Schafer A, Pfeiffer P, Vielmetter W. Cell. 1990;60:921–928. doi: 10.1016/0092-8674(90)90340-k. [DOI] [PubMed] [Google Scholar]
- 42.Clark J M. Gene. 1991;104:75–80. doi: 10.1016/0378-1119(91)90467-p. [DOI] [PubMed] [Google Scholar]
- 43.King J S, Fairley C F, Morgan W F. J Biol Chem. 1994;269:13061–13064. [PubMed] [Google Scholar]
- 44.King J S, Fairley C F, Morgan W F. J Biol Chem. 1996;271:20450–20457. doi: 10.1074/jbc.271.34.20450. [DOI] [PubMed] [Google Scholar]
- 45.Clark J M, Joyce C M, Beardsley G P. J Mol Biol. 1987;198:123–127. doi: 10.1016/0022-2836(87)90462-1. [DOI] [PubMed] [Google Scholar]
- 46.Clark J M. Nucleic Acids Res. 1988;16:9677–9686. doi: 10.1093/nar/16.20.9677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Beese L S, Derbyshire V, Steitz T A. Science. 1993;260:352–355. doi: 10.1126/science.8469987. [DOI] [PubMed] [Google Scholar]