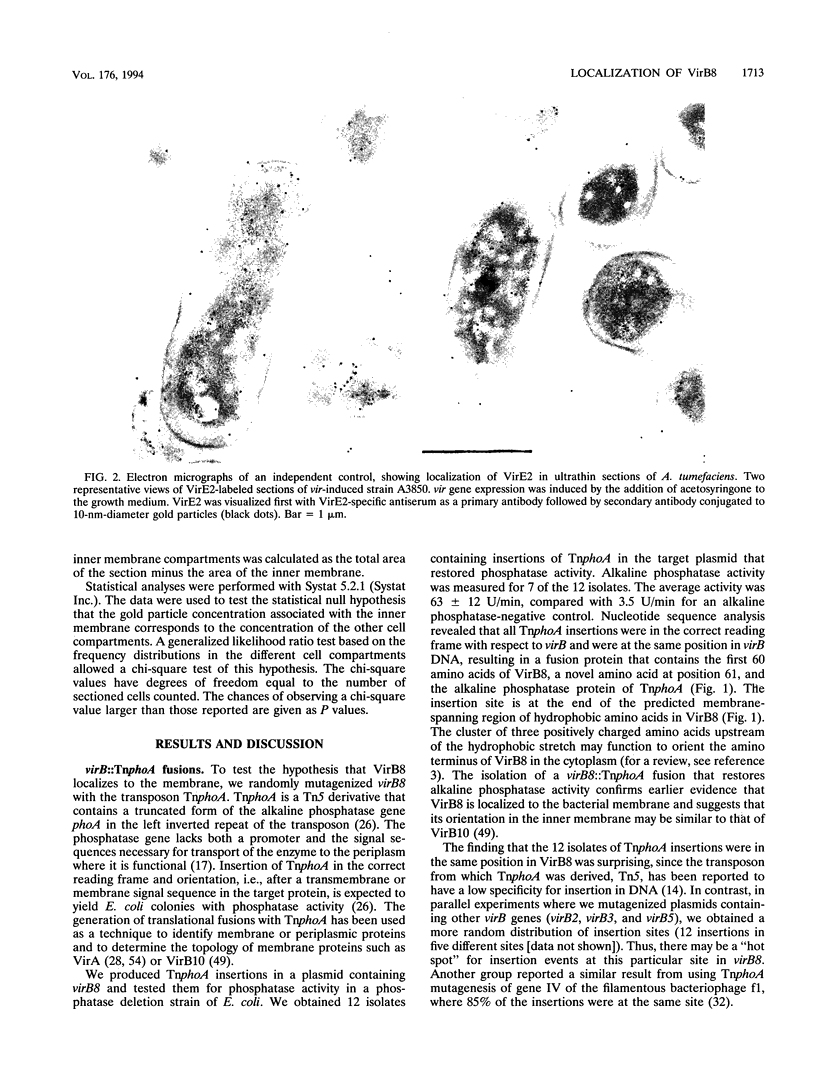
Abstract
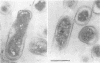
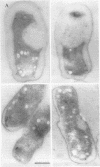
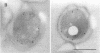
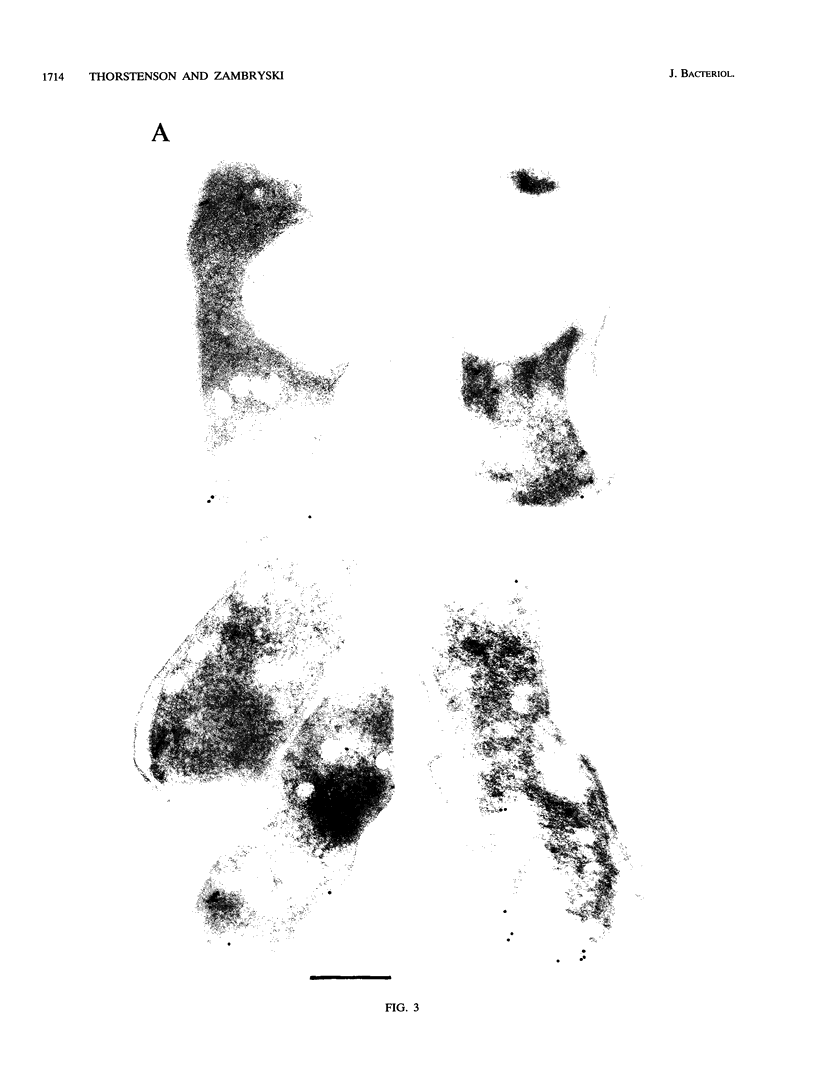
Agrobacterium tumefaciens genetically transforms plant cells by transferring a specific DNA fragment from the bacterium through several biological membranes to the plant nucleus where the DNA is integrated. This complex DNA transport process likely involves membrane-localized proteins in both the plant and the bacterium. The 11 hydrophobic or membrane-localized proteins of the virB operon are excellent candidates to have a role in DNA export from agrobacteria. Here, we show by TnphoA mutagenesis and immunogold electron microscopy that one of the VirB proteins, VirB8, is located at the inner membrane. The observation that a virB8::TnphoA fusion restores export of alkaline phosphatase to the periplasm suggests that VirB8 spans the inner membrane. Immunogold labeling of VirB8 was detected on the inner membrane of vir-induced A. tumefaciens by transmission electron microscopy. Compared with that of the controls, VirB8 labeling was significantly greater on the inner membrane than on the other cell compartments. These results confirm the inner membrane localization of VirB8 and strengthen the hypothesis that VirB proteins help form a transfer DNA export channel or gate.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alley M. R., Maddock J. R., Shapiro L. Polar localization of a bacterial chemoreceptor. Genes Dev. 1992 May;6(5):825–836. doi: 10.1101/gad.6.5.825. [DOI] [PubMed] [Google Scholar]
- Berger B. R., Christie P. J. The Agrobacterium tumefaciens virB4 gene product is an essential virulence protein requiring an intact nucleoside triphosphate-binding domain. J Bacteriol. 1993 Mar;175(6):1723–1734. doi: 10.1128/jb.175.6.1723-1734.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd D., Beckwith J. The role of charged amino acids in the localization of secreted and membrane proteins. Cell. 1990 Sep 21;62(6):1031–1033. doi: 10.1016/0092-8674(90)90378-r. [DOI] [PubMed] [Google Scholar]
- Brickman E., Beckwith J. Analysis of the regulation of Escherichia coli alkaline phosphatase synthesis using deletions and phi80 transducing phages. J Mol Biol. 1975 Aug 5;96(2):307–316. doi: 10.1016/0022-2836(75)90350-2. [DOI] [PubMed] [Google Scholar]
- Chen E. Y., Seeburg P. H. Supercoil sequencing: a fast and simple method for sequencing plasmid DNA. DNA. 1985 Apr;4(2):165–170. doi: 10.1089/dna.1985.4.165. [DOI] [PubMed] [Google Scholar]
- Christie P. J., Ward J. E., Jr, Gordon M. P., Nester E. W. A gene required for transfer of T-DNA to plants encodes an ATPase with autophosphorylating activity. Proc Natl Acad Sci U S A. 1989 Dec;86(24):9677–9681. doi: 10.1073/pnas.86.24.9677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie P. J., Ward J. E., Winans S. C., Nester E. W. The Agrobacterium tumefaciens virE2 gene product is a single-stranded-DNA-binding protein that associates with T-DNA. J Bacteriol. 1988 Jun;170(6):2659–2667. doi: 10.1128/jb.170.6.2659-2667.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citovsky V., DE Vos G., Zambryski P. Single-Stranded DNA Binding Protein Encoded by the virE Locus of Agrobacterium tumefaciens. Science. 1988 Apr 22;240(4851):501–504. doi: 10.1126/science.240.4851.501. [DOI] [PubMed] [Google Scholar]
- Citovsky V., Zupan J., Warnick D., Zambryski P. Nuclear localization of Agrobacterium VirE2 protein in plant cells. Science. 1992 Jun 26;256(5065):1802–1805. doi: 10.1126/science.1615325. [DOI] [PubMed] [Google Scholar]
- Cook D. M., Farrand S. K. The oriT region of the Agrobacterium tumefaciens Ti plasmid pTiC58 shares DNA sequence identity with the transfer origins of RSF1010 and RK2/RP4 and with T-region borders. J Bacteriol. 1992 Oct;174(19):6238–6246. doi: 10.1128/jb.174.19.6238-6246.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covacci A., Rappuoli R. Pertussis toxin export requires accessory genes located downstream from the pertussis toxin operon. Mol Microbiol. 1993 May;8(3):429–434. doi: 10.1111/j.1365-2958.1993.tb01587.x. [DOI] [PubMed] [Google Scholar]
- Dale E. M., Binns A. N., Ward J. E., Jr Construction and characterization of Tn5virB, a transposon that generates nonpolar mutations, and its use to define virB8 as an essential virulence gene in Agrobacterium tumefaciens. J Bacteriol. 1993 Feb;175(3):887–891. doi: 10.1128/jb.175.3.887-891.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A. Agrobacterium tumefaciens virE operon encodes a single-stranded DNA-binding protein. Proc Natl Acad Sci U S A. 1988 May;85(9):2909–2913. doi: 10.1073/pnas.85.9.2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engström P., Zambryski P., Van Montagu M., Stachel S. Characterization of Agrobacterium tumefaciens virulence proteins induced by the plant factor acetosyringone. J Mol Biol. 1987 Oct 20;197(4):635–645. doi: 10.1016/0022-2836(87)90470-0. [DOI] [PubMed] [Google Scholar]
- Gutierrez C., Barondess J., Manoil C., Beckwith J. The use of transposon TnphoA to detect genes for cell envelope proteins subject to a common regulatory stimulus. Analysis of osmotically regulated genes in Escherichia coli. J Mol Biol. 1987 May 20;195(2):289–297. doi: 10.1016/0022-2836(87)90650-4. [DOI] [PubMed] [Google Scholar]
- Hoffman C. S., Wright A. Fusions of secreted proteins to alkaline phosphatase: an approach for studying protein secretion. Proc Natl Acad Sci U S A. 1985 Aug;82(15):5107–5111. doi: 10.1073/pnas.82.15.5107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuldau G. A., De Vos G., Owen J., McCaffrey G., Zambryski P. The virB operon of Agrobacterium tumefaciens pTiC58 encodes 11 open reading frames. Mol Gen Genet. 1990 Apr;221(2):256–266. doi: 10.1007/BF00261729. [DOI] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Lessl M., Balzer D., Pansegrau W., Lanka E. Sequence similarities between the RP4 Tra2 and the Ti VirB region strongly support the conjugation model for T-DNA transfer. J Biol Chem. 1992 Oct 5;267(28):20471–20480. [PubMed] [Google Scholar]
- Maddock J. R., Shapiro L. Polar location of the chemoreceptor complex in the Escherichia coli cell. Science. 1993 Mar 19;259(5102):1717–1723. doi: 10.1126/science.8456299. [DOI] [PubMed] [Google Scholar]
- Manoil C., Beckwith J. TnphoA: a transposon probe for protein export signals. Proc Natl Acad Sci U S A. 1985 Dec;82(23):8129–8133. doi: 10.1073/pnas.82.23.8129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthysse A. G., Holmes K. V., Gurlitz R. H. Elaboration of cellulose fibrils by Agrobacterium tumefaciens during attachment to carrot cells. J Bacteriol. 1981 Jan;145(1):583–595. doi: 10.1128/jb.145.1.583-595.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melchers L. S., Regensburg-Tuïnk T. J., Bourret R. B., Sedee N. J., Schilperoort R. A., Hooykaas P. J. Membrane topology and functional analysis of the sensory protein VirA of Agrobacterium tumefaciens. EMBO J. 1989 Jul;8(7):1919–1925. doi: 10.1002/j.1460-2075.1989.tb03595.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto S., Toyoda-Yamamoto A., Ito K., Takebe I., Machida Y. Localization and orientation of the VirD4 protein of Agrobacterium tumefaciens in the cell membrane. Mol Gen Genet. 1991 Aug;228(1-2):24–32. doi: 10.1007/BF00282443. [DOI] [PubMed] [Google Scholar]
- Pansegrau W., Lanka E. Common sequence motifs in DNA relaxases and nick regions from a variety of DNA transfer systems. Nucleic Acids Res. 1991 Jun 25;19(12):3455–3455. doi: 10.1093/nar/19.12.3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pansegrau W., Ziegelin G., Lanka E. Covalent association of the traI gene product of plasmid RP4 with the 5'-terminal nucleotide at the relaxation nick site. J Biol Chem. 1990 Jun 25;265(18):10637–10644. [PubMed] [Google Scholar]
- Russel M., Kaźmierczak B. Analysis of the structure and subcellular location of filamentous phage pIV. J Bacteriol. 1993 Jul;175(13):3998–4007. doi: 10.1128/jb.175.13.3998-4007.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirasu K., Kado C. I. Membrane location of the Ti plasmid VirB proteins involved in the biosynthesis of a pilin-like conjugative structure on Agrobacterium tumefaciens. FEMS Microbiol Lett. 1993 Aug 1;111(2-3):287–294. doi: 10.1111/j.1574-6968.1993.tb06400.x. [DOI] [PubMed] [Google Scholar]
- Shirasu K., Kado C. I. The virB operon of the Agrobacterium tumefaciens virulence regulon has sequence similarities to B, C and D open reading frames downstream of the pertussis toxin-operon and to the DNA transfer-operons of broad-host-range conjugative plasmids. Nucleic Acids Res. 1993 Jan 25;21(2):353–354. doi: 10.1093/nar/21.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirasu K., Morel P., Kado C. I. Characterization of the virB operon of an Agrobacterium tumefaciens Ti plasmid: nucleotide sequence and protein analysis. Mol Microbiol. 1990 Jul;4(7):1153–1163. doi: 10.1111/j.1365-2958.1990.tb00690.x. [DOI] [PubMed] [Google Scholar]
- Stachel S. E., An G., Flores C., Nester E. W. A Tn3 lacZ transposon for the random generation of beta-galactosidase gene fusions: application to the analysis of gene expression in Agrobacterium. EMBO J. 1985 Apr;4(4):891–898. doi: 10.1002/j.1460-2075.1985.tb03715.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stachel S. E., Nester E. W. The genetic and transcriptional organization of the vir region of the A6 Ti plasmid of Agrobacterium tumefaciens. EMBO J. 1986 Jul;5(7):1445–1454. doi: 10.1002/j.1460-2075.1986.tb04381.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stachel S. E., Zambryski P. C. Agrobacterium tumefaciens and the susceptible plant cell: a novel adaptation of extracellular recognition and DNA conjugation. Cell. 1986 Oct 24;47(2):155–157. doi: 10.1016/0092-8674(86)90437-x. [DOI] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci U S A. 1985 Feb;82(4):1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson D. V., Melchers L. S., Idler K. B., Schilperoort R. A., Hooykaas P. J. Analysis of the complete nucleotide sequence of the Agrobacterium tumefaciens virB operon. Nucleic Acids Res. 1988 May 25;16(10):4621–4636. doi: 10.1093/nar/16.10.4621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorstenson Y. R., Kuldau G. A., Zambryski P. C. Subcellular localization of seven VirB proteins of Agrobacterium tumefaciens: implications for the formation of a T-DNA transport structure. J Bacteriol. 1993 Aug;175(16):5233–5241. doi: 10.1128/jb.175.16.5233-5241.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vestweber D., Schatz G. DNA-protein conjugates can enter mitochondria via the protein import pathway. Nature. 1989 Mar 9;338(6211):170–172. doi: 10.1038/338170a0. [DOI] [PubMed] [Google Scholar]
- Ward J. E., Akiyoshi D. E., Regier D., Datta A., Gordon M. P., Nester E. W. Characterization of the virB operon from an Agrobacterium tumefaciens Ti plasmid. J Biol Chem. 1988 Apr 25;263(12):5804–5814. [PubMed] [Google Scholar]
- Ward J. E., Akiyoshi D. E., Regier D., Datta A., Gordon M. P., Nester E. W. Correction: characterization of the virB operon from Agrobacterium tumefaciens Ti plasmid. J Biol Chem. 1990 Mar 15;265(8):4768–4768. [PubMed] [Google Scholar]
- Ward J. E., Jr, Dale E. M., Christie P. J., Nester E. W., Binns A. N. Complementation analysis of Agrobacterium tumefaciens Ti plasmid virB genes by use of a vir promoter expression vector: virB9, virB10, and virB11 are essential virulence genes. J Bacteriol. 1990 Sep;172(9):5187–5199. doi: 10.1128/jb.172.9.5187-5199.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward J. E., Jr, Dale E. M., Nester E. W., Binns A. N. Identification of a virB10 protein aggregate in the inner membrane of Agrobacterium tumefaciens. J Bacteriol. 1990 Sep;172(9):5200–5210. doi: 10.1128/jb.172.9.5200-5210.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters V. L., Hirata K. H., Pansegrau W., Lanka E., Guiney D. G. Sequence identity in the nick regions of IncP plasmid transfer origins and T-DNA borders of Agrobacterium Ti plasmids. Proc Natl Acad Sci U S A. 1991 Feb 15;88(4):1456–1460. doi: 10.1073/pnas.88.4.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss A. A., Johnson F. D., Burns D. L. Molecular characterization of an operon required for pertussis toxin secretion. Proc Natl Acad Sci U S A. 1993 Apr 1;90(7):2970–2974. doi: 10.1073/pnas.90.7.2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winans S. C., Kerstetter R. A., Ward J. E., Nester E. W. A protein required for transcriptional regulation of Agrobacterium virulence genes spans the cytoplasmic membrane. J Bacteriol. 1989 Mar;171(3):1616–1622. doi: 10.1128/jb.171.3.1616-1622.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winans S. C. Two-way chemical signaling in Agrobacterium-plant interactions. Microbiol Rev. 1992 Mar;56(1):12–31. doi: 10.1128/mr.56.1.12-31.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambryski P., Joos H., Genetello C., Leemans J., Montagu M. V., Schell J. Ti plasmid vector for the introduction of DNA into plant cells without alteration of their normal regeneration capacity. EMBO J. 1983;2(12):2143–2150. doi: 10.1002/j.1460-2075.1983.tb01715.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegelin G., Pansegrau W., Strack B., Balzer D., Kröger M., Kruft V., Lanka E. Nucleotide sequence and organization of genes flanking the transfer origin of promiscuous plasmid RP4. DNA Seq. 1991;1(5):303–327. doi: 10.3109/10425179109020786. [DOI] [PubMed] [Google Scholar]
- Zorick T. S., Schooley C. Ultrarapid freezing on a diamond surface. Microsc Res Tech. 1992 Jan 1;20(1):103–104. doi: 10.1002/jemt.1070200112. [DOI] [PubMed] [Google Scholar]
- de Bruijn F. J., Lupski J. R. The use of transposon Tn5 mutagenesis in the rapid generation of correlated physical and genetic maps of DNA segments cloned into multicopy plasmids--a review. Gene. 1984 Feb;27(2):131–149. doi: 10.1016/0378-1119(84)90135-5. [DOI] [PubMed] [Google Scholar]