Abstract
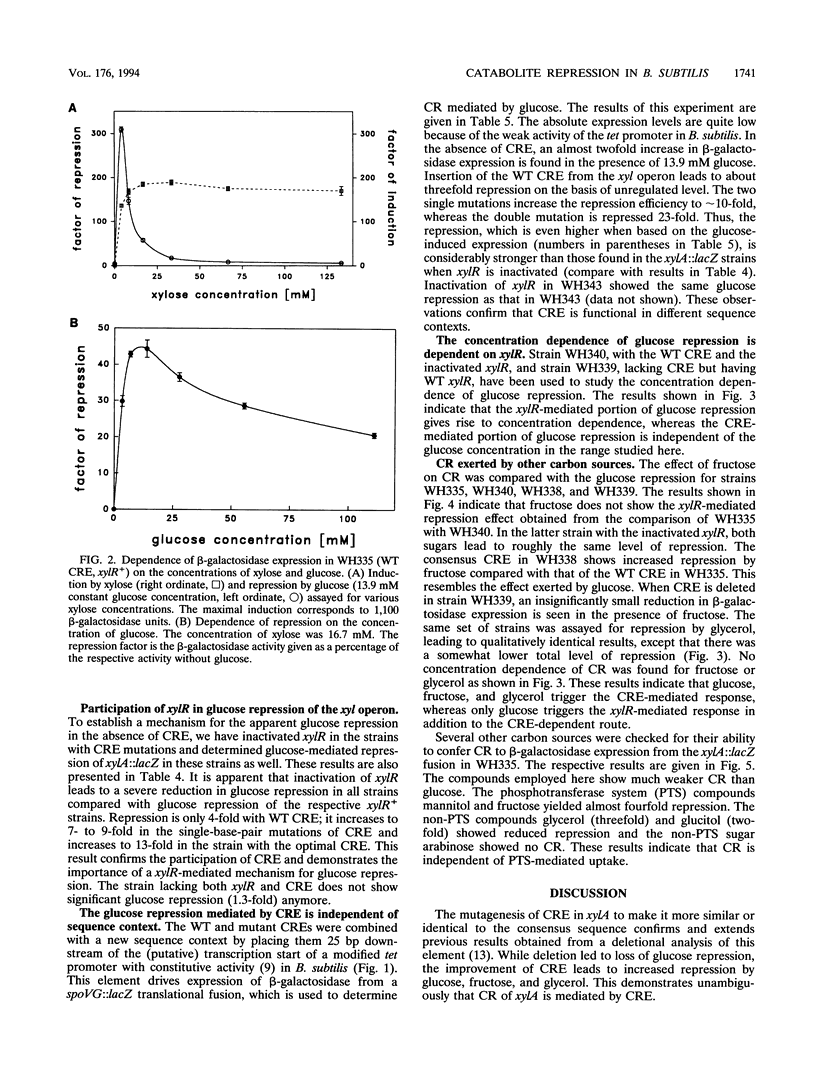
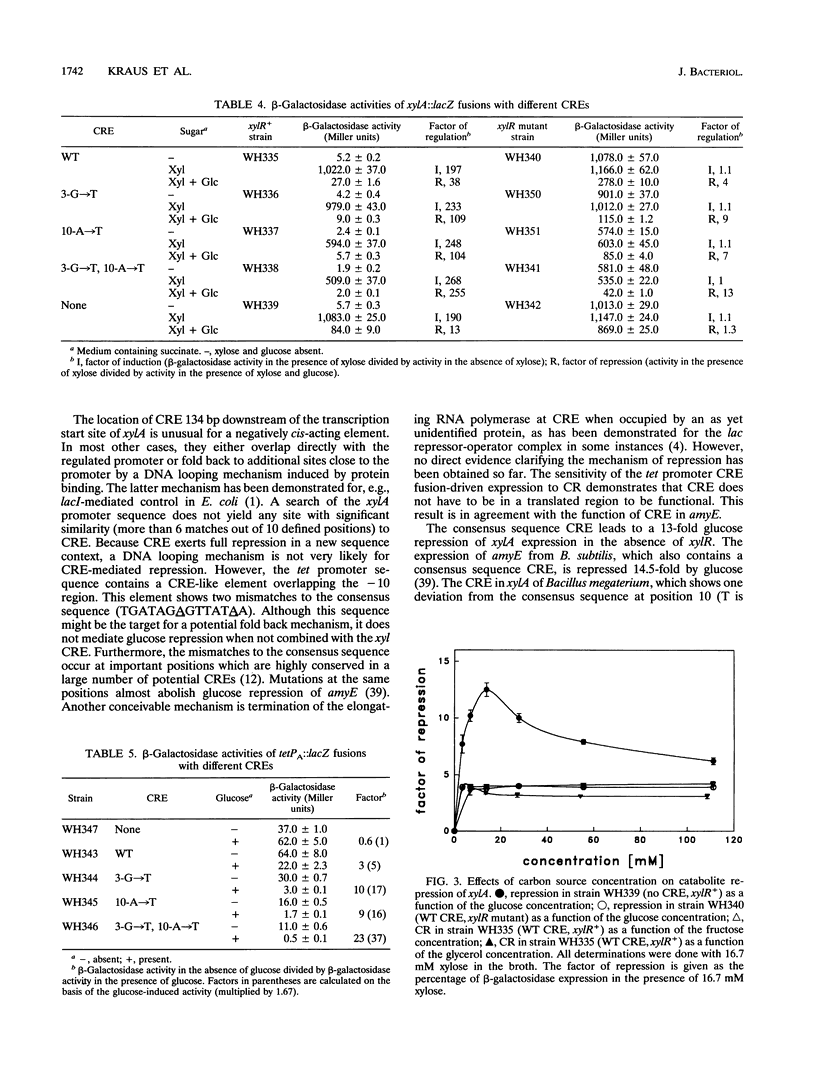
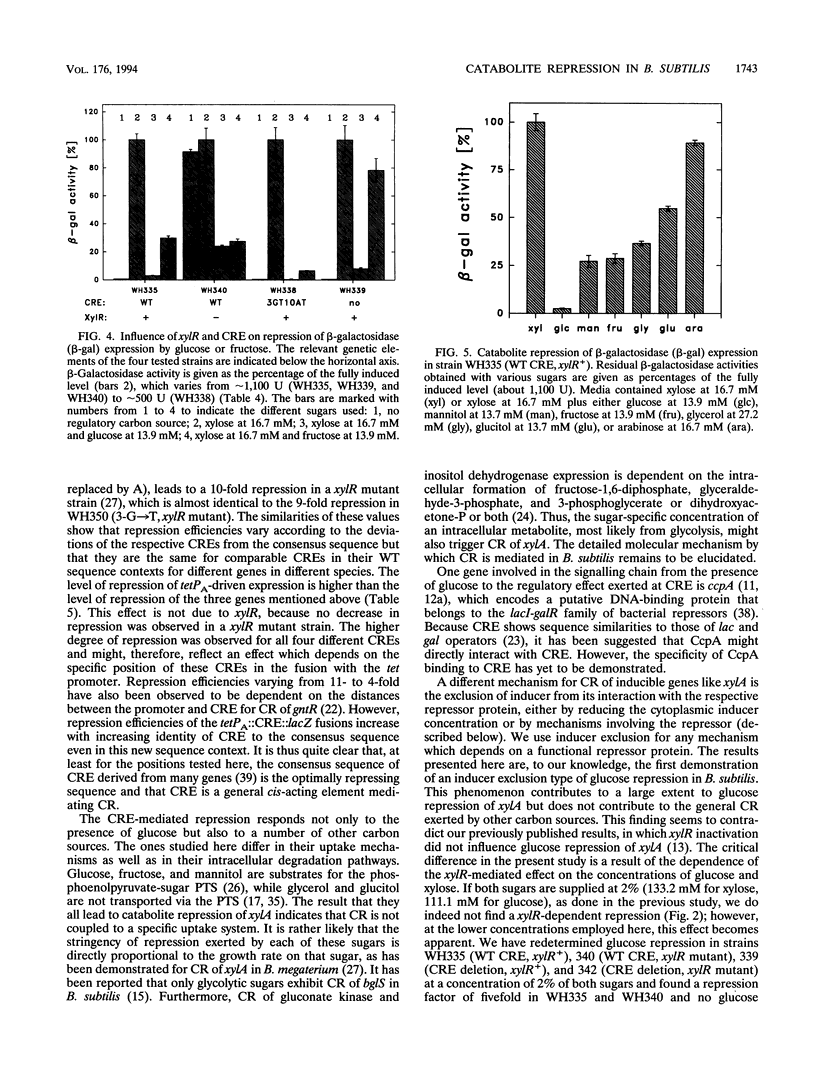
Catabolite repression (CR) of xylose utilization by Bacillus subtilis involves a 14-bp cis-acting element (CRE) located in the translated region of the gene encoding xylose isomerase (xylA). Mutations of CRE making it more similar to a previously proposed consensus element lead to increased CR exerted by glucose, fructose, and glycerol. Fusion of CRE to an unrelated, constitutive promoter confers CR to beta-galactosidase expression directed by that promoter. This result demonstrates that CRE can function independently of sequence context and suggests that it is indeed a generally active cis element for CR. In contrast to the other carbon sources studied here, glucose leads to an additional repression of xylA expression, which is independent of CRE and is not found when CRE is fused to the unrelated promoter. This repression requires a functional xylR encoding Xyl repressor and is dependent on the concentrations of glucose and the inducer xylose in the culture broth. Potential mechanisms for this glucose-specific repression are discussed.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Chakerian A. E., Matthews K. S. Effect of lac repressor oligomerization on regulatory outcome. Mol Microbiol. 1992 Apr;6(8):963–968. doi: 10.1111/j.1365-2958.1992.tb02162.x. [DOI] [PubMed] [Google Scholar]
- Dekker K., Yamagata H., Sakaguchi K., Udaka S. Xylose (glucose) isomerase gene from the thermophile Thermus thermophilus: cloning, sequencing, and comparison with other thermostable xylose isomerases. J Bacteriol. 1991 May;173(10):3078–3083. doi: 10.1128/jb.173.10.3078-3083.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deuschle U., Gentz R., Bujard H. lac Repressor blocks transcribing RNA polymerase and terminates transcription. Proc Natl Acad Sci U S A. 1986 Jun;83(12):4134–4137. doi: 10.1073/pnas.83.12.4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher S. H., Sonenshein A. L. Control of carbon and nitrogen metabolism in Bacillus subtilis. Annu Rev Microbiol. 1991;45:107–135. doi: 10.1146/annurev.mi.45.100191.000543. [DOI] [PubMed] [Google Scholar]
- Fouet A., Jin S. F., Raffel G., Sonenshein A. L. Multiple regulatory sites in the Bacillus subtilis citB promoter region. J Bacteriol. 1990 Sep;172(9):5408–5415. doi: 10.1128/jb.172.9.5408-5415.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissendörfer M., Hillen W. Regulated expression of heterologous genes in Bacillus subtilis using the Tn10 encoded tet regulatory elements. Appl Microbiol Biotechnol. 1990 Sep;33(6):657–663. doi: 10.1007/BF00604933. [DOI] [PubMed] [Google Scholar]
- Gärtner D., Degenkolb J., Ripperger J. A., Allmansberger R., Hillen W. Regulation of the Bacillus subtilis W23 xylose utilization operon: interaction of the Xyl repressor with the xyl operator and the inducer xylose. Mol Gen Genet. 1992 Apr;232(3):415–422. doi: 10.1007/BF00266245. [DOI] [PubMed] [Google Scholar]
- Gärtner D., Geissendörfer M., Hillen W. Expression of the Bacillus subtilis xyl operon is repressed at the level of transcription and is induced by xylose. J Bacteriol. 1988 Jul;170(7):3102–3109. doi: 10.1128/jb.170.7.3102-3109.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henkin T. M., Grundy F. J., Nicholson W. L., Chambliss G. H. Catabolite repression of alpha-amylase gene expression in Bacillus subtilis involves a trans-acting gene product homologous to the Escherichia coli lacl and galR repressors. Mol Microbiol. 1991 Mar;5(3):575–584. doi: 10.1111/j.1365-2958.1991.tb00728.x. [DOI] [PubMed] [Google Scholar]
- Jacob S., Allmansberger R., Gärtner D., Hillen W. Catabolite repression of the operon for xylose utilization from Bacillus subtilis W23 is mediated at the level of transcription and depends on a cis site in the xylA reading frame. Mol Gen Genet. 1991 Oct;229(2):189–196. doi: 10.1007/BF00272155. [DOI] [PubMed] [Google Scholar]
- Kreuzer P., Gärtner D., Allmansberger R., Hillen W. Identification and sequence analysis of the Bacillus subtilis W23 xylR gene and xyl operator. J Bacteriol. 1989 Jul;171(7):3840–3845. doi: 10.1128/jb.171.7.3840-3845.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krüger S., Stülke J., Hecker M. Catabolite repression of beta-glucanase synthesis in Bacillus subtilis. J Gen Microbiol. 1993 Sep;139(9):2047–2054. doi: 10.1099/00221287-139-9-2047. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Lopez J. M., Thoms B. Role of sugar uptake and metabolic intermediates on catabolite repression in Bacillus subtilis. J Bacteriol. 1977 Jan;129(1):217–224. doi: 10.1128/jb.129.1.217-224.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J., Vieira J. A new pair of M13 vectors for selecting either DNA strand of double-digest restriction fragments. Gene. 1982 Oct;19(3):269–276. doi: 10.1016/0378-1119(82)90016-6. [DOI] [PubMed] [Google Scholar]
- Miwa Y., Fujita Y. Determination of the cis sequence involved in catabolite repression of the Bacillus subtilis gnt operon; implication of a consensus sequence in catabolite repression in the genus Bacillus. Nucleic Acids Res. 1990 Dec 11;18(23):7049–7053. doi: 10.1093/nar/18.23.7049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miwa Y., Fujita Y. Promoter-independent catabolite repression of the Bacillus subtilis gnt operon. J Biochem. 1993 Jun;113(6):665–671. doi: 10.1093/oxfordjournals.jbchem.a124100. [DOI] [PubMed] [Google Scholar]
- Nicholson W. L., Park Y. K., Henkin T. M., Won M., Weickert M. J., Gaskell J. A., Chambliss G. H. Catabolite repression-resistant mutations of the Bacillus subtilis alpha-amylase promoter affect transcription levels and are in an operator-like sequence. J Mol Biol. 1987 Dec 20;198(4):609–618. doi: 10.1016/0022-2836(87)90204-x. [DOI] [PubMed] [Google Scholar]
- Nihashi J., Fujita Y. Catabolite repression of inositol dehydrogenase and gluconate kinase syntheses in Bacillus subtilis. Biochim Biophys Acta. 1984 Mar 22;798(1):88–95. doi: 10.1016/0304-4165(84)90014-x. [DOI] [PubMed] [Google Scholar]
- Oda M., Katagai T., Tomura D., Shoun H., Hoshino T., Furukawa K. Analysis of the transcriptional activity of the hut promoter in Bacillus subtilis and identification of a cis-acting regulatory region associated with catabolite repression downstream from the site of transcription. Mol Microbiol. 1992 Sep;6(18):2573–2582. doi: 10.1111/j.1365-2958.1992.tb01434.x. [DOI] [PubMed] [Google Scholar]
- Reizer J., Saier M. H., Jr, Deutscher J., Grenier F., Thompson J., Hengstenberg W. The phosphoenolpyruvate:sugar phosphotransferase system in gram-positive bacteria: properties, mechanism, and regulation. Crit Rev Microbiol. 1988;15(4):297–338. doi: 10.3109/10408418809104461. [DOI] [PubMed] [Google Scholar]
- Rygus T., Hillen W. Catabolite repression of the xyl operon in Bacillus megaterium. J Bacteriol. 1992 May;174(9):3049–3055. doi: 10.1128/jb.174.9.3049-3055.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saier M. H., Jr A multiplicity of potential carbon catabolite repression mechanisms in prokaryotic and eukaryotic microorganisms. New Biol. 1991 Dec;3(12):1137–1147. [PubMed] [Google Scholar]
- Saier M. H., Jr Regulatory interactions involving the proteins of the phosphotransferase system in enteric bacteria. J Cell Biochem. 1993 Jan;51(1):62–68. doi: 10.1002/jcb.240510112. [DOI] [PubMed] [Google Scholar]
- Shimotsu H., Henner D. J. Construction of a single-copy integration vector and its use in analysis of regulation of the trp operon of Bacillus subtilis. Gene. 1986;43(1-2):85–94. doi: 10.1016/0378-1119(86)90011-9. [DOI] [PubMed] [Google Scholar]
- Sizemore C., Geissdörfer W., Hillen W. Using fusions with luxAB from Vibrio harveyi MAV to quantify induction and catabolite repression of the xyl operon in Staphylococcus carnosus TM300. FEMS Microbiol Lett. 1993 Mar 1;107(2-3):303–306. doi: 10.1111/j.1574-6968.1993.tb06047.x. [DOI] [PubMed] [Google Scholar]
- Sizemore C., Wieland B., Götz F., Hillen W. Regulation of Staphylococcus xylosus xylose utilization genes at the molecular level. J Bacteriol. 1992 May;174(9):3042–3048. doi: 10.1128/jb.174.9.3042-3048.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart G. C. Catabolite repression in the gram-positive bacteria: generation of negative regulators of transcription. J Cell Biochem. 1993 Jan;51(1):25–28. doi: 10.1002/jcb.240510106. [DOI] [PubMed] [Google Scholar]
- Tangney M., Priest F. G., Mitchell W. J. Two glucose transport systems in Bacillus licheniformis. J Bacteriol. 1993 Apr;175(7):2137–2142. doi: 10.1128/jb.175.7.2137-2142.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weickert M. J., Adhya S. A family of bacterial regulators homologous to Gal and Lac repressors. J Biol Chem. 1992 Aug 5;267(22):15869–15874. [PubMed] [Google Scholar]
- Weickert M. J., Chambliss G. H. Site-directed mutagenesis of a catabolite repression operator sequence in Bacillus subtilis. Proc Natl Acad Sci U S A. 1990 Aug;87(16):6238–6242. doi: 10.1073/pnas.87.16.6238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm M., Hollenberg C. P. Selective cloning of Bacillus subtilis xylose isomerase and xylulokinase in Escherichia coli genes by IS5-mediated expression. EMBO J. 1984 Nov;3(11):2555–2560. doi: 10.1002/j.1460-2075.1984.tb02173.x. [DOI] [PMC free article] [PubMed] [Google Scholar]


