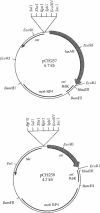
Abstract
Gene fusions are frequently used to facilitate studies of gene expression and promoter activity. We have found that certain reporter genes can, themselves, influence promoter activity. For example, the commonly used luxAB reporter genes can activate or repress transcription from a subset of promoters, generating data apparently at odds with those obtained with other reporter genes. These effects are probably related to an intrinsically curved DNA segment in the 5' coding sequence of the luxA gene. Thus, caution must be observed when one is interpreting results obtained with a single reporter gene system such as luxAB.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Casadaban M. J. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. J Mol Biol. 1976 Jul 5;104(3):541–555. doi: 10.1016/0022-2836(76)90119-4. [DOI] [PubMed] [Google Scholar]
- Close T. J., Rodriguez R. L. Construction and characterization of the chloramphenicol-resistance gene cartridge: a new approach to the transcriptional mapping of extrachromosomal elements. Gene. 1982 Dec;20(2):305–316. doi: 10.1016/0378-1119(82)90048-8. [DOI] [PubMed] [Google Scholar]
- Dattananda C. S., Rajkumari K., Gowrishankar J. Multiple mechanisms contribute to osmotic inducibility of proU operon expression in Escherichia coli: demonstration of two osmoresponsive promoters and of a negative regulatory element within the first structural gene. J Bacteriol. 1991 Dec;173(23):7481–7490. doi: 10.1128/jb.173.23.7481-7490.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlap V. J., Csonka L. N. Osmotic regulation of L-proline transport in Salmonella typhimurium. J Bacteriol. 1985 Jul;163(1):296–304. doi: 10.1128/jb.163.1.296-304.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engebrecht J., Simon M., Silverman M. Measuring gene expression with light. Science. 1985 Mar 15;227(4692):1345–1347. doi: 10.1126/science.2983423. [DOI] [PubMed] [Google Scholar]
- Gemmill R. M., Jones J. W., Haughn G. W., Calvo J. M. Transcription initiation sites of the leucine operons of Salmonella typhimurium and Escherichia coli. J Mol Biol. 1983 Oct 15;170(1):39–59. doi: 10.1016/s0022-2836(83)80226-5. [DOI] [PubMed] [Google Scholar]
- Gemmill R. M., Tripp M., Friedman S. B., Calvo J. M. Promoter mutation causing catabolite repression of the Salmonella typhimurium leucine operon. J Bacteriol. 1984 Jun;158(3):948–953. doi: 10.1128/jb.158.3.948-953.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins C. F., Dorman C. J., Stirling D. A., Waddell L., Booth I. R., May G., Bremer E. A physiological role for DNA supercoiling in the osmotic regulation of gene expression in S. typhimurium and E. coli. Cell. 1988 Feb 26;52(4):569–584. doi: 10.1016/0092-8674(88)90470-9. [DOI] [PubMed] [Google Scholar]
- Hinton J. C., Santos D. S., Seirafi A., Hulton C. S., Pavitt G. D., Higgins C. F. Expression and mutational analysis of the nucleoid-associated protein H-NS of Salmonella typhimurium. Mol Microbiol. 1992 Aug;6(16):2327–2337. doi: 10.1111/j.1365-2958.1992.tb01408.x. [DOI] [PubMed] [Google Scholar]
- Lenny A. B., Margolin P. Locations of the opp and supX genes of Salmonella typhimurium and Escherichia coli. J Bacteriol. 1980 Aug;143(2):747–752. doi: 10.1128/jb.143.2.747-752.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley D. M., Higgins C. F. Local DNA topology and gene expression: the case of the leu-500 promoter. Mol Microbiol. 1991 Apr;5(4):779–783. doi: 10.1111/j.1365-2958.1991.tb00749.x. [DOI] [PubMed] [Google Scholar]
- Margolin P., Zumstein L., Sternglanz R., Wang J. C. The Escherichia coli supX locus is topA, the structural gene for DNA topoisomerase I. Proc Natl Acad Sci U S A. 1985 Aug;82(16):5437–5441. doi: 10.1073/pnas.82.16.5437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenney K., Shimatake H., Court D., Schmeissner U., Brady C., Rosenberg M. A system to study promoter and terminator signals recognized by Escherichia coli RNA polymerase. Gene Amplif Anal. 1981;2:383–415. [PubMed] [Google Scholar]
- Menzel R., Gellert M. Fusions of the Escherichia coli gyrA and gyrB control regions to the galactokinase gene are inducible by coumermycin treatment. J Bacteriol. 1987 Mar;169(3):1272–1278. doi: 10.1128/jb.169.3.1272-1278.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller V. L., Mekalanos J. J. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol. 1988 Jun;170(6):2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norqvist A., Wolf-Watz H. Characterization of a novel chromosomal virulence locus involved in expression of a major surface flagellar sheath antigen of the fish pathogen Vibrio anguillarum. Infect Immun. 1993 Jun;61(6):2434–2444. doi: 10.1128/iai.61.6.2434-2444.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson O., Koncz C., Szalay A. A. The use of the luxA gene of the bacterial luciferase operon as a reporter gene. Mol Gen Genet. 1988 Dec;215(1):1–9. doi: 10.1007/BF00331295. [DOI] [PubMed] [Google Scholar]
- Overdier D. G., Csonka L. N. A transcriptional silencer downstream of the promoter in the osmotically controlled proU operon of Salmonella typhimurium. Proc Natl Acad Sci U S A. 1992 Apr 1;89(7):3140–3144. doi: 10.1073/pnas.89.7.3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen-Hughes T. A., Pavitt G. D., Santos D. S., Sidebotham J. M., Hulton C. S., Hinton J. C., Higgins C. F. The chromatin-associated protein H-NS interacts with curved DNA to influence DNA topology and gene expression. Cell. 1992 Oct 16;71(2):255–265. doi: 10.1016/0092-8674(92)90354-f. [DOI] [PubMed] [Google Scholar]
- Park S. F., Stirling D. A., Hulton C. S., Booth I. R., Higgins C. F., Stewart G. S. A novel, non-invasive promoter probe vector: cloning of the osmoregulated proU promoter of Escherichia coli K12. Mol Microbiol. 1989 Aug;3(8):1011–1023. doi: 10.1111/j.1365-2958.1989.tb00252.x. [DOI] [PubMed] [Google Scholar]
- Richardson S. M., Higgins C. F., Lilley D. M. DNA supercoiling and the leu-500 promoter mutation of Salmonella typhimurium. EMBO J. 1988 Jun;7(6):1863–1869. doi: 10.1002/j.1460-2075.1988.tb03019.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimpiläinen M., Forsberg A., Wolf-Watz H. A novel protein, LcrQ, involved in the low-calcium response of Yersinia pseudotuberculosis shows extensive homology to YopH. J Bacteriol. 1992 May;174(10):3355–3363. doi: 10.1128/jb.174.10.3355-3363.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Searles L. L., Wessler S. R., Calvo J. M. Transcription attenuation is the major mechanism by which the leu operon of Salmonella typhimurium is controlled. J Mol Biol. 1983 Jan 25;163(3):377–394. doi: 10.1016/0022-2836(83)90064-5. [DOI] [PubMed] [Google Scholar]
- Shaw W. V. Chloramphenicol acetyltransferase from chloramphenicol-resistant bacteria. Methods Enzymol. 1975;43:737–755. doi: 10.1016/0076-6879(75)43141-x. [DOI] [PubMed] [Google Scholar]
- Silhavy T. J., Beckwith J. R. Uses of lac fusions for the study of biological problems. Microbiol Rev. 1985 Dec;49(4):398–418. doi: 10.1128/mr.49.4.398-418.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons R. W., Houman F., Kleckner N. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene. 1987;53(1):85–96. doi: 10.1016/0378-1119(87)90095-3. [DOI] [PubMed] [Google Scholar]
- Stirling D. A., Hulton C. S., Waddell L., Park S. F., Stewart G. S., Booth I. R., Higgins C. F. Molecular characterization of the proU loci of Salmonella typhimurium and Escherichia coli encoding osmoregulated glycine betaine transport systems. Mol Microbiol. 1989 Aug;3(8):1025–1038. doi: 10.1111/j.1365-2958.1989.tb00253.x. [DOI] [PubMed] [Google Scholar]
- Takeshita S., Sato M., Toba M., Masahashi W., Hashimoto-Gotoh T. High-copy-number and low-copy-number plasmid vectors for lacZ alpha-complementation and chloramphenicol- or kanamycin-resistance selection. Gene. 1987;61(1):63–74. doi: 10.1016/0378-1119(87)90365-9. [DOI] [PubMed] [Google Scholar]
- Thein S. L., Hinton J. A simple and rapid method of direct sequencing using Dynabeads. Br J Haematol. 1991 Sep;79(1):113–115. doi: 10.1111/j.1365-2141.1991.tb08016.x. [DOI] [PubMed] [Google Scholar]
- Tupper A. E., Owen-Hughes T. A., Ussery D. W., Santos D. S., Ferguson D. J., Sidebotham J. M., Hinton J. C., Higgins C. F. The chromatin-associated protein H-NS alters DNA topology in vitro. EMBO J. 1994 Jan 1;13(1):258–268. doi: 10.1002/j.1460-2075.1994.tb06256.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lorenzo V., Eltis L., Kessler B., Timmis K. N. Analysis of Pseudomonas gene products using lacIq/Ptrp-lac plasmids and transposons that confer conditional phenotypes. Gene. 1993 Jan 15;123(1):17–24. doi: 10.1016/0378-1119(93)90533-9. [DOI] [PubMed] [Google Scholar]