Abstract
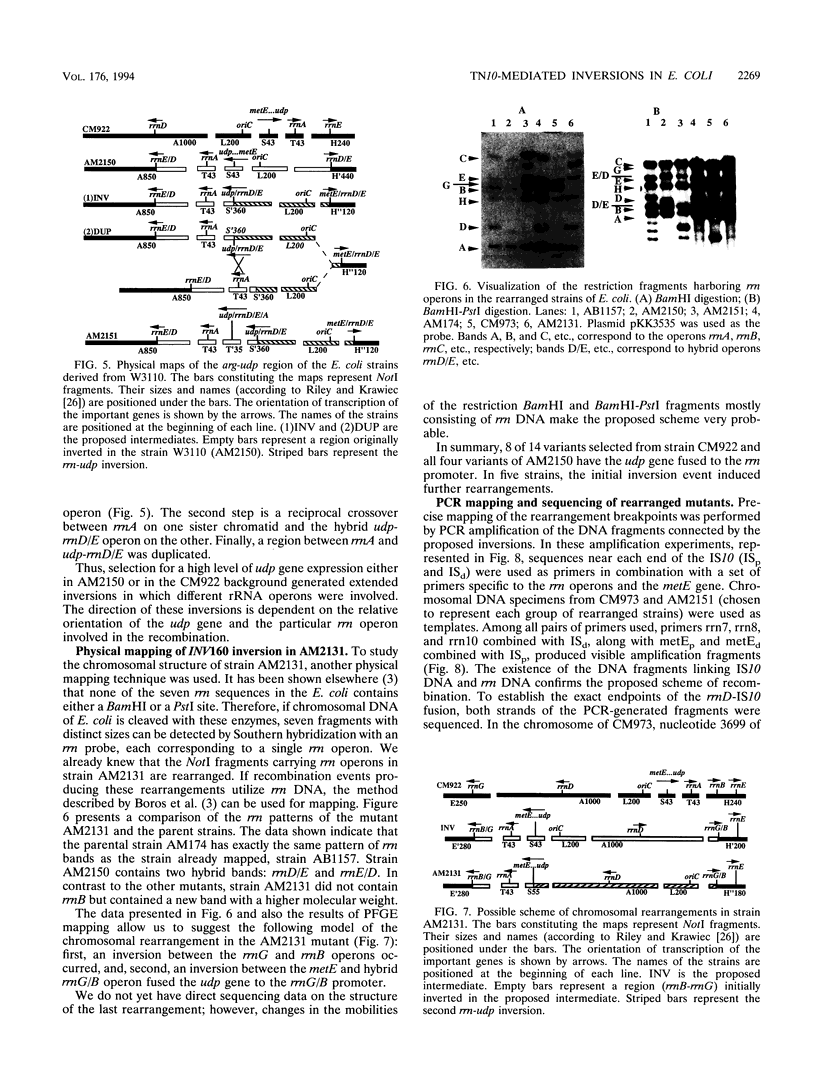
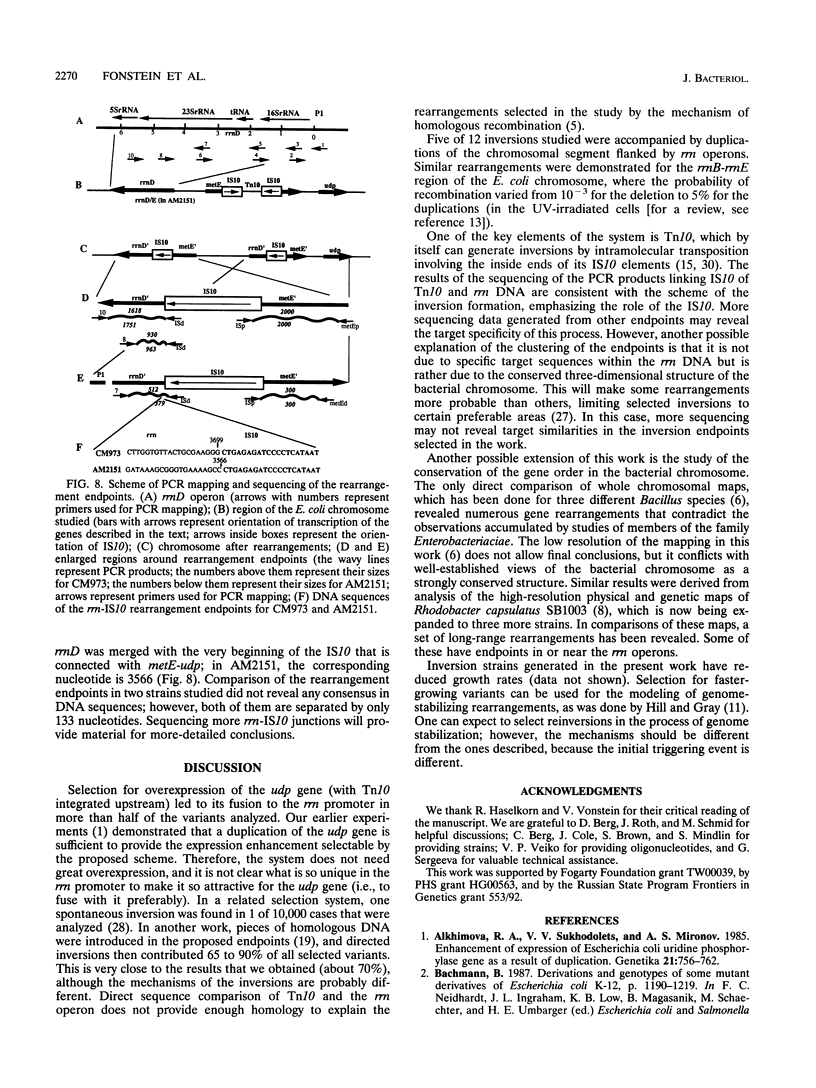
Two strains carrying metE::Tn10 insertions (upstream of the udp gene) were used to isolate mutants of Escherichia coli overexpressing udp. These strains differ in their gene order; one contains an inversion between the rrnD and rrnE rRNA operons. Selection was based on the ability of overexpressed Udp to complement thymine auxotrophy. Chromosomal rearrangements that connect the udp gene and promoters of different rrn operons were obtained by this selection. Seven of 14 independent mutants selected in one of the initial strains contained similar inversions of the metE-rrnD segment of the chromosome (about 12% of its length). Another mutant contained traces of a more complicated event, inversion between rrnB and rrnG operons, which was followed by reinversion of the segment between metE and the hybrid rrnG/B operon. Similar inversions (udp-rrn) in a strain already carrying an rrnE-rrnD inversion flip the chromosomal segment between metE and rrnD/E in the opposite direction. In this case, inversions are also accompanied by duplications of the chromosomal region between the rrnA and hybrid udp-rrnD/E operons. PCR amplification with a set of oligonucleotides from the rrn, Tn5, and met genes was used for more detailed mapping. Amplified fragments of the rearranged chromosomes connecting rrnD sequences and insertion elements were sequenced, and inversion endpoints were established.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alkhimova R. A., Sukhodolets V. V., Mironov A. S. Usilenie ekspressii gena uridinfosforilazy (udp) Escherichia coli v rezul'tate duplikatsii. Genetika. 1985 May;21(5):756–762. [PubMed] [Google Scholar]
- Boros I., Kiss A., Venetianer P. Physical map of the seven ribosomal RNA genes of Escherichia coli. Nucleic Acids Res. 1979;6(5):1817–1830. doi: 10.1093/nar/6.5.1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brikun I. A., Mironov A. S., Masiliunaite R. V., Sukhodolets V. V. Klonirovanie gena uridinfosforilazy Escherichia coli: lokalizatsiia strukturnoi i reguliatornoi oblastei gena na klonirovannom fragmentei identifikatsiia belkovogo produkta. Mol Gen Mikrobiol Virusol. 1990 Jun;(6):7–11. [PubMed] [Google Scholar]
- Brosius J., Dull T. J., Sleeter D. D., Noller H. F. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J Mol Biol. 1981 May 15;148(2):107–127. doi: 10.1016/0022-2836(81)90508-8. [DOI] [PubMed] [Google Scholar]
- Carlson C. R., Grønstad A., Kolstø A. B. Physical maps of the genomes of three Bacillus cereus strains. J Bacteriol. 1992 Jun;174(11):3750–3756. doi: 10.1128/jb.174.11.3750-3756.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonstein M., Haselkorn R. Chromosomal structure of Rhodobacter capsulatus strain SB1003: cosmid encyclopedia and high-resolution physical and genetic map. Proc Natl Acad Sci U S A. 1993 Mar 15;90(6):2522–2526. doi: 10.1073/pnas.90.6.2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonstein M., Zheng S., Haselkorn R. Physical map of the genome of Rhodobacter capsulatus SB 1003. J Bacteriol. 1992 Jun;174(12):4070–4077. doi: 10.1128/jb.174.12.4070-4077.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman P. E., Hartman Z., Stahl R. C. Classification and mapping of spontaneous and induced mutations in the histidine operon of Salmonella. Adv Genet. 1971;16:1–34. doi: 10.1016/s0065-2660(08)60352-1. [DOI] [PubMed] [Google Scholar]
- Hill C. W., Gray J. A. Effects of chromosomal inversion on cell fitness in Escherichia coli K-12. Genetics. 1988 Aug;119(4):771–778. doi: 10.1093/genetics/119.4.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill C. W., Harnish B. W. Inversions between ribosomal RNA genes of Escherichia coli. Proc Natl Acad Sci U S A. 1981 Nov;78(11):7069–7072. doi: 10.1073/pnas.78.11.7069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingston R. E., Gutell R. R., Taylor A. R., Chamberlin M. J. Transcriptional mapping of plasmid pKK3535. Quantitation of the effect of guanosine tetraphosphate on binding to the rrnB promoters and a lambda promoter with sequence homologies in the CII binding region. J Mol Biol. 1981 Mar 15;146(4):433–449. doi: 10.1016/0022-2836(81)90041-3. [DOI] [PubMed] [Google Scholar]
- Kleckner N., Reichardt K., Botstein D. Inversions and deletions of the Salmonella chromosome generated by the translocatable tetracycline resistance element Tn10. J Mol Biol. 1979 Jan 5;127(1):89–115. doi: 10.1016/0022-2836(79)90461-3. [DOI] [PubMed] [Google Scholar]
- Konrad E. B. Method for the isolation of Escherichia coli mutants with enhanced recombination between chromosomal duplications. J Bacteriol. 1977 Apr;130(1):167–172. doi: 10.1128/jb.130.1.167-172.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulakauskas S. T., Sukhodolets V. V., Mironov A. S. Inversiia khromosomy, soprovozhdaiushchaiasia usileniem ékspressii gena uridinfosforilazy u Escherichia coli K-12. Genetika. 1985 Mar;21(3):375–383. [PubMed] [Google Scholar]
- Mironov A. S., Sukhodolets V. V. Promoter-like mutants with increased expression of the Escherichia coli uridine phosphorylase structural gene. J Bacteriol. 1979 Feb;137(2):802–810. doi: 10.1128/jb.137.2.802-810.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray V., Martin R. F. The degree of ultraviolet light damage to DNA containing iododeoxyuridine or bromodeoxyuridine is dependent on the DNA sequence. Nucleic Acids Res. 1989 Apr 11;17(7):2675–2691. doi: 10.1093/nar/17.7.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebollo J. E., François V., Louarn J. M. Detection and possible role of two large nondivisible zones on the Escherichia coli chromosome. Proc Natl Acad Sci U S A. 1988 Dec;85(24):9391–9395. doi: 10.1073/pnas.85.24.9391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley M., Anilionis A. Evolution of the bacterial genome. Annu Rev Microbiol. 1978;32:519–560. doi: 10.1146/annurev.mi.32.100178.002511. [DOI] [PubMed] [Google Scholar]
- Schmid M. B., Roth J. R. Genetic methods for analysis and manipulation of inversion mutations in bacteria. Genetics. 1983 Nov;105(3):517–537. doi: 10.1093/genetics/105.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segall A., Mahan M. J., Roth J. R. Rearrangement of the bacterial chromosome: forbidden inversions. Science. 1988 Sep 9;241(4871):1314–1318. doi: 10.1126/science.3045970. [DOI] [PubMed] [Google Scholar]
- Shen M. M., Raleigh E. A., Kleckner N. Physical analysis of Tn10- and IS10-promoted transpositions and rearrangements. Genetics. 1987 Jul;116(3):359–369. doi: 10.1093/genetics/116.3.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C. L., Econome J. G., Schutt A., Klco S., Cantor C. R. A physical map of the Escherichia coli K12 genome. Science. 1987 Jun 12;236(4807):1448–1453. doi: 10.1126/science.3296194. [DOI] [PubMed] [Google Scholar]
- Smith C. L., Kolodner R. D. Mapping of Escherichia coli chromosomal Tn5 and F insertions by pulsed field gel electrophoresis. Genetics. 1988 Jun;119(2):227–236. doi: 10.1093/genetics/119.2.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young K. K., Edlin G. J., Wilson G. G. Genetic analysis of bacteriophage T4 transducing bacteriophages. J Virol. 1982 Jan;41(1):345–347. doi: 10.1128/jvi.41.1.345-347.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]