Abstract
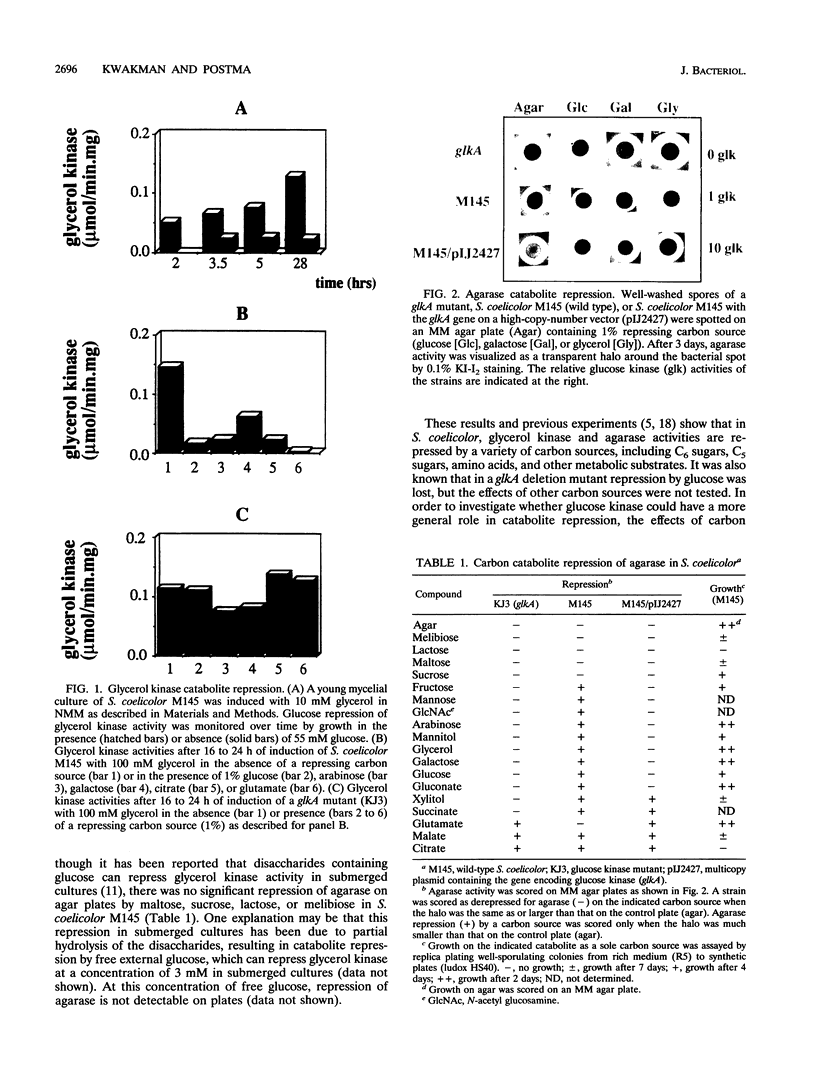
A glucose kinase (glkA) mutant of Streptomyces coelicolor A3(2) M145 was selected by the ability to grow in the presence of the nonmetabolizable glucose analog 2-deoxyglucose. In this glkA mutant, carbon catabolite repression of glycerol kinase and agarase was relieved on several carbon sources tested, even though most of these carbon sources are not metabolized via glucose kinase. This suggests that catabolite repression is not regulated by the flux through glucose kinase and that the protein itself has a regulatory role in carbon catabolite repression. A 10-fold overproduction of glucose kinase also results in relief of catabolite repression, suggesting that excess glucose kinase can titrate the repressing signal away. This could be achieved directly by competition of excess glucose kinase with its repressing form for binding sites on DNA promoter regions or indirectly by competition for binding of another regulatory protein.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Angell S., Schwarz E., Bibb M. J. The glucose kinase gene of Streptomyces coelicolor A3(2): its nucleotide sequence, transcriptional analysis and role in glucose repression. Mol Microbiol. 1992 Oct;6(19):2833–2844. doi: 10.1111/j.1365-2958.1992.tb01463.x. [DOI] [PubMed] [Google Scholar]
- Ayer D. E., Kretzner L., Eisenman R. N. Mad: a heterodimeric partner for Max that antagonizes Myc transcriptional activity. Cell. 1993 Jan 29;72(2):211–222. doi: 10.1016/0092-8674(93)90661-9. [DOI] [PubMed] [Google Scholar]
- Bennett W. S., Jr, Steitz T. A. Glucose-induced conformational change in yeast hexokinase. Proc Natl Acad Sci U S A. 1978 Oct;75(10):4848–4852. doi: 10.1073/pnas.75.10.4848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibb M. J., Jones G. H., Joseph R., Buttner M. J., Ward J. M. The agarase gene (dag A) of Streptomyces coelicolor A3(2): affinity purification and characterization of the cloned gene product. J Gen Microbiol. 1987 Aug;133(8):2089–2096. doi: 10.1099/00221287-133-8-2089. [DOI] [PubMed] [Google Scholar]
- Duckett C. S., Perkins N. D., Kowalik T. F., Schmid R. M., Huang E. S., Baldwin A. S., Jr, Nabel G. J. Dimerization of NF-KB2 with RelA(p65) regulates DNA binding, transcriptional activation, and inhibition by an I kappa B-alpha (MAD-3). Mol Cell Biol. 1993 Mar;13(3):1315–1322. doi: 10.1128/mcb.13.3.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gancedo J. M. Carbon catabolite repression in yeast. Eur J Biochem. 1992 Jun 1;206(2):297–313. doi: 10.1111/j.1432-1033.1992.tb16928.x. [DOI] [PubMed] [Google Scholar]
- Hoggett J. G., Kellett G. L. Kinetics of the monomer-dimer reaction of yeast hexokinase PI. Biochem J. 1992 Oct 15;287(Pt 2):567–572. doi: 10.1042/bj2870567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda H., Seno E. T., Bruton C. J., Chater K. F. Genetic mapping, cloning and physiological aspects of the glucose kinase gene of Streptomyces coelicolor. Mol Gen Genet. 1984;196(3):501–507. doi: 10.1007/BF00436199. [DOI] [PubMed] [Google Scholar]
- Kreuzer P., Gärtner D., Allmansberger R., Hillen W. Identification and sequence analysis of the Bacillus subtilis W23 xylR gene and xyl operator. J Bacteriol. 1989 Jul;171(7):3840–3845. doi: 10.1128/jb.171.7.3840-3845.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H., Bloom L. M., Walsh C. T., Botstein D. The residual enzymatic phosphorylation activity of hexokinase II mutants is correlated with glucose repression in Saccharomyces cerevisiae. Mol Cell Biol. 1989 Dec;9(12):5643–5649. doi: 10.1128/mcb.9.12.5643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plumbridge J. A. Sequence of the nagBACD operon in Escherichia coli K12 and pattern of transcription within the nag regulon. Mol Microbiol. 1989 Apr;3(4):505–515. doi: 10.1111/j.1365-2958.1989.tb00197.x. [DOI] [PubMed] [Google Scholar]
- Postma P. W., Lengeler J. W., Jacobson G. R. Phosphoenolpyruvate:carbohydrate phosphotransferase systems of bacteria. Microbiol Rev. 1993 Sep;57(3):543–594. doi: 10.1128/mr.57.3.543-594.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose M., Albig W., Entian K. D. Glucose repression in Saccharomyces cerevisiae is directly associated with hexose phosphorylation by hexokinases PI and PII. Eur J Biochem. 1991 Aug 1;199(3):511–518. doi: 10.1111/j.1432-1033.1991.tb16149.x. [DOI] [PubMed] [Google Scholar]
- Sauer F., Jäckle H. Concentration-dependent transcriptional activation or repression by Krüppel from a single binding site. Nature. 1991 Oct 10;353(6344):563–566. doi: 10.1038/353563a0. [DOI] [PubMed] [Google Scholar]
- Seno E. T., Chater K. F. Glycerol catabolic enzymes and their regulation in wild-type and mutant strains of Streptomyces coelicolor A3(2). J Gen Microbiol. 1983 May;129(5):1403–1413. doi: 10.1099/00221287-129-5-1403. [DOI] [PubMed] [Google Scholar]
- Smith P. K., Krohn R. I., Hermanson G. T., Mallia A. K., Gartner F. H., Provenzano M. D., Fujimoto E. K., Goeke N. M., Olson B. J., Klenk D. C. Measurement of protein using bicinchoninic acid. Anal Biochem. 1985 Oct;150(1):76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- Solenberg P. J., Baltz R. H. Transposition of Tn5096 and other IS493 derivatives in Streptomyces griseofuscus. J Bacteriol. 1991 Feb;173(3):1096–1104. doi: 10.1128/jb.173.3.1096-1104.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trumbly R. J. Glucose repression in the yeast Saccharomyces cerevisiae. Mol Microbiol. 1992 Jan;6(1):15–21. doi: 10.1111/j.1365-2958.1992.tb00832.x. [DOI] [PubMed] [Google Scholar]



