Abstract
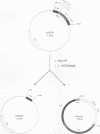
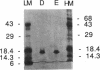
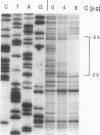
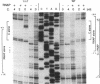
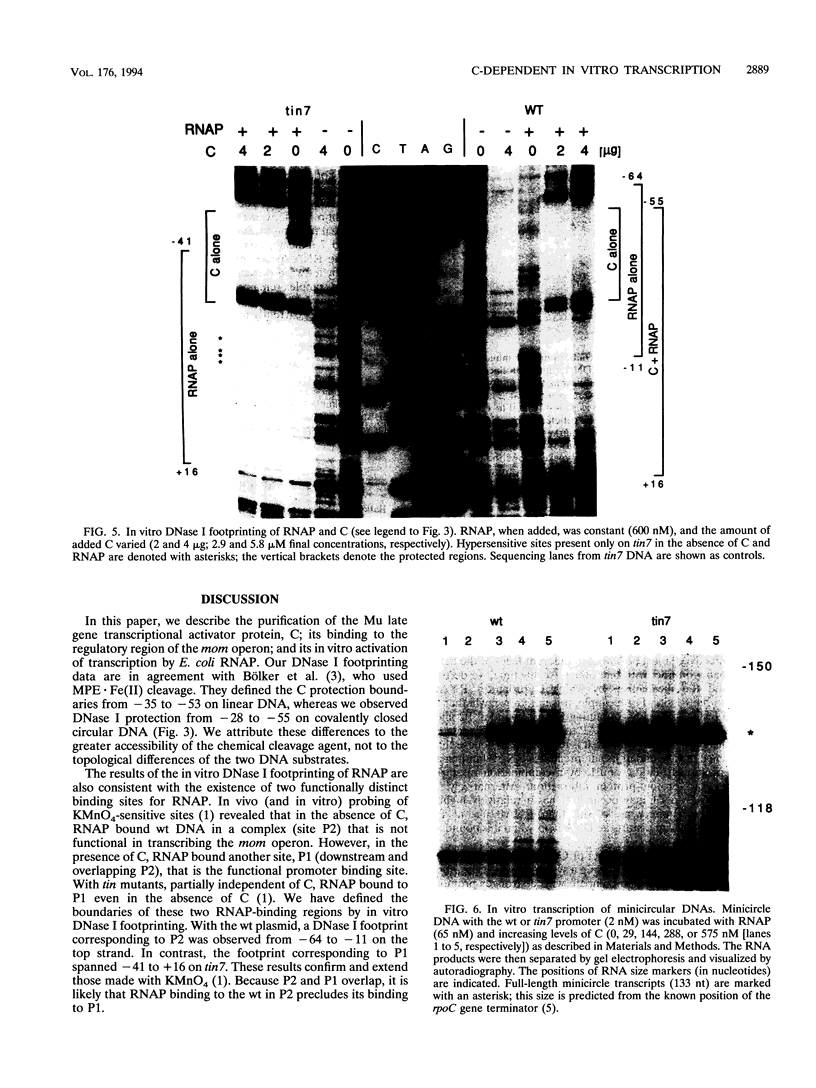
The phage Mu gene C encodes a 16.5-kDa site-specific DNA-binding protein that functions as a trans-activator of the four phage "late" operons, including mom. We have overexpressed and purified C and used it for DNase I footprinting and transcription analyses in vitro. The footprinting results are summarized as follows. (i) As shown previously (V. Balke, V. Nagaraja, T. Gindlesperger, and S. Hattman, Nucleic Acids Res. 12:2777-2784, 1992) in vivo, Escherichia coli RNA polymerase (RNAP) bound the wild-type (wt) mom promoter at a site slightly upstream from the functionally active site bound on the C-independent tin7 mutant promoter. (ii) In the presence of C, however, RNAP bound the wt promoter at the same site as tin7. (iii) C and RNAP were both bound by the mom promoter at overlapping sites, indicating that they were probably on different faces of the DNA helix. The minicircle system of Choy and Adhya (H. E. Choy and S. Adhya, Proc. Natl. Acad. Sci. USA 90:472-476, 1993) was used to compare transcription in vitro from the wt and tin7 promoters. This analysis showed the following. (i) Few full-length transcripts were observed from the wt promoter in the absence of C, but addition of increasing amounts of C greatly stimulated transcription. (ii) RNA was transcribed from the tin7 promoter in the absence of C, but addition of C had a small stimulatory effect. (iii) Transcription from linearized minicircles or restriction fragment templates was greatly reduced (although still stimulated by C) with both the wt and tin7 promoters. These results show that C alone is capable of activating rightward transcription in vitro by promoting RNAP binding at a functionally active site. Additionally, DNA topology plays an important role in transcriptional activation in vitro.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Balke V., Nagaraja V., Gindlesperger T., Hattman S. Functionally distinct RNA polymerase binding sites in the phage Mu mom promoter region. Nucleic Acids Res. 1992 Jun 11;20(11):2777–2784. doi: 10.1093/nar/20.11.2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bölker M., Kahmann R. The Escherichia coli regulatory protein OxyR discriminates between methylated and unmethylated states of the phage Mu mom promoter. EMBO J. 1989 Aug;8(8):2403–2410. doi: 10.1002/j.1460-2075.1989.tb08370.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bölker M., Wulczyn F. G., Kahmann R. Role of bacteriophage Mu C protein in activation of the mom gene promoter. J Bacteriol. 1989 Apr;171(4):2019–2027. doi: 10.1128/jb.171.4.2019-2027.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadaban M. J., Chou J., Cohen S. N. In vitro gene fusions that join an enzymatically active beta-galactosidase segment to amino-terminal fragments of exogenous proteins: Escherichia coli plasmid vectors for the detection and cloning of translational initiation signals. J Bacteriol. 1980 Aug;143(2):971–980. doi: 10.1128/jb.143.2.971-980.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy H. E., Adhya S. RNA polymerase idling and clearance in gal promoters: use of supercoiled minicircle DNA template made in vivo. Proc Natl Acad Sci U S A. 1993 Jan 15;90(2):472–476. doi: 10.1073/pnas.90.2.472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattman S., Brooks J. E., Masurekar M. Sequence specificity of the P1 modification methylase (M.Eco P1) and the DNA methylase (M.Eco dam) controlled by the Escherichia coli dam gene. J Mol Biol. 1978 Dec 15;126(3):367–380. doi: 10.1016/0022-2836(78)90046-3. [DOI] [PubMed] [Google Scholar]
- Hattman S. DNA methyltransferase-dependent transcription of the phage Mu mom gene. Proc Natl Acad Sci U S A. 1982 Sep;79(18):5518–5521. doi: 10.1073/pnas.79.18.5518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattman S., Ives J., Margolin W., Howe M. M. Regulation and expression of the bacteriophage mu mom gene: mapping of the transactivation (dad) function to the C region. Gene. 1985;39(1):71–76. doi: 10.1016/0378-1119(85)90109-x. [DOI] [PubMed] [Google Scholar]
- Hattman S., Ives J. S1 nuclease mapping of the phage Mu mom gene promoter: a model for the regulation of mom expression. Gene. 1984 Jul-Aug;29(1-2):185–198. doi: 10.1016/0378-1119(84)90179-3. [DOI] [PubMed] [Google Scholar]
- Hattman S., Ives J., Wall L., Marić S. The bacteriophage Mu com gene appears to specify a translation factor required for mom gene expression. Gene. 1987;55(2-3):345–351. doi: 10.1016/0378-1119(87)90295-2. [DOI] [PubMed] [Google Scholar]
- Hattman S. Unusual modification of bacteriophage Mu DNA. J Virol. 1979 Nov;32(2):468–475. doi: 10.1128/jvi.32.2.468-475.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heisig P., Kahmann R. The sequence and mom-transactivation function of the C gene of bacteriophage Mu. Gene. 1986;43(1-2):59–67. doi: 10.1016/0378-1119(86)90008-9. [DOI] [PubMed] [Google Scholar]
- Kahmann R. Methylation regulates the expression of a DNA-modification function encoded by bacteriophage Mu. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 2):639–646. doi: 10.1101/sqb.1983.047.01.075. [DOI] [PubMed] [Google Scholar]
- Kahmann R. Methylation regulates the expression of a DNA-modification function encoded by bacteriophage Mu. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 2):639–646. doi: 10.1101/sqb.1983.047.01.075. [DOI] [PubMed] [Google Scholar]
- Kahmann R., Seiler A., Wulczyn F. G., Pfaff E. The mom gene of bacteriophage mu: a unique regulatory scheme to control a lethal function. Gene. 1985;39(1):61–70. doi: 10.1016/0378-1119(85)90108-8. [DOI] [PubMed] [Google Scholar]
- Margolin W., Howe M. M. Activation of the bacteriophage Mu lys promoter by Mu C protein requires the sigma 70 subunit of Escherichia coli RNA polymerase. J Bacteriol. 1990 Mar;172(3):1424–1429. doi: 10.1128/jb.172.3.1424-1429.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolin W., Howe M. M. Localization and DNA sequence analysis of the C gene of bacteriophage Mu, the positive regulator of Mu late transcription. Nucleic Acids Res. 1986 Jun 25;14(12):4881–4897. doi: 10.1093/nar/14.12.4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolin W., Rao G., Howe M. M. Bacteriophage Mu late promoters: four late transcripts initiate near a conserved sequence. J Bacteriol. 1989 Apr;171(4):2003–2018. doi: 10.1128/jb.171.4.2003-2018.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaraja V., Hecht G., Hattman S. The phage Mu 'late' gene transcription activator, C, is a site-specific DNA binding protein. Biochem Pharmacol. 1988 May 1;37(9):1809–1810. doi: 10.1016/0006-2952(88)90457-1. [DOI] [PubMed] [Google Scholar]
- Plasterk R. H., Vrieling H., Van de Putte P. Transcription initiation of Mu mom depends on methylation of the promoter region and a phage-coded transactivator. Nature. 1983 Jan 27;301(5898):344–347. doi: 10.1038/301344a0. [DOI] [PubMed] [Google Scholar]
- Sasse-Dwight S., Gralla J. D. KMnO4 as a probe for lac promoter DNA melting and mechanism in vivo. J Biol Chem. 1989 May 15;264(14):8074–8081. [PubMed] [Google Scholar]
- Seiler A., Blöcker H., Frank R., Kahmann R. The mom gene of bacteriophage Mu: the mechanism of methylation-dependent expression. EMBO J. 1986 Oct;5(10):2719–2728. doi: 10.1002/j.1460-2075.1986.tb04556.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swinton D., Hattman S., Crain P. F., Cheng C. S., Smith D. L., McCloskey J. A. Purification and characterization of the unusual deoxynucleoside, alpha-N-(9-beta-D-2'-deoxyribofuranosylpurin-6-yl)glycinamide, specified by the phage Mu modification function. Proc Natl Acad Sci U S A. 1983 Dec;80(24):7400–7404. doi: 10.1073/pnas.80.24.7400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toussaint A. DNA modification of bacteriophage Mu-1 requires both host and bacteriophage functions. J Virol. 1977 Sep;23(3):825–826. doi: 10.1128/jvi.23.3.825-826.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toussaint A. The DNA modification function of temperate phage Mu-1. Virology. 1976 Mar;70(1):17–27. doi: 10.1016/0042-6822(76)90232-4. [DOI] [PubMed] [Google Scholar]
- Wulczyn F. G., Bölker M., Kahmann R. Translation of the bacteriophage Mu mom gene is positively regulated by the phage com gene product. Cell. 1989 Jun 30;57(7):1201–1210. doi: 10.1016/0092-8674(89)90057-3. [DOI] [PubMed] [Google Scholar]
- Wulczyn F. G., Kahmann R. Post-transcriptional regulation of the bacteriophage Mu mom gene by the com gene product. Gene. 1987;51(2-3):139–147. doi: 10.1016/0378-1119(87)90302-7. [DOI] [PubMed] [Google Scholar]
- Wulczyn F. G., Kahmann R. Translational stimulation: RNA sequence and structure requirements for binding of Com protein. Cell. 1991 Apr 19;65(2):259–269. doi: 10.1016/0092-8674(91)90160-z. [DOI] [PubMed] [Google Scholar]
- Zengel J. M., Mueckl D., Lindahl L. Protein L4 of the E. coli ribosome regulates an eleven gene r protein operon. Cell. 1980 Sep;21(2):523–535. doi: 10.1016/0092-8674(80)90490-0. [DOI] [PubMed] [Google Scholar]