Abstract
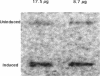
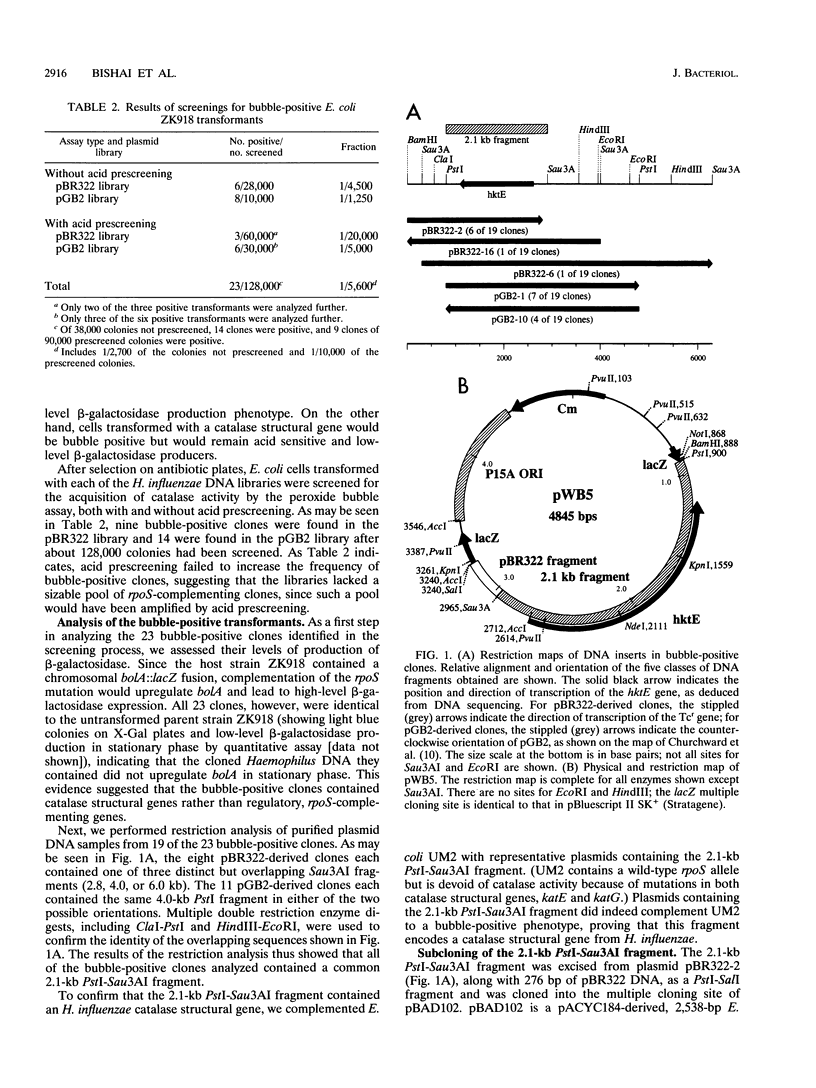
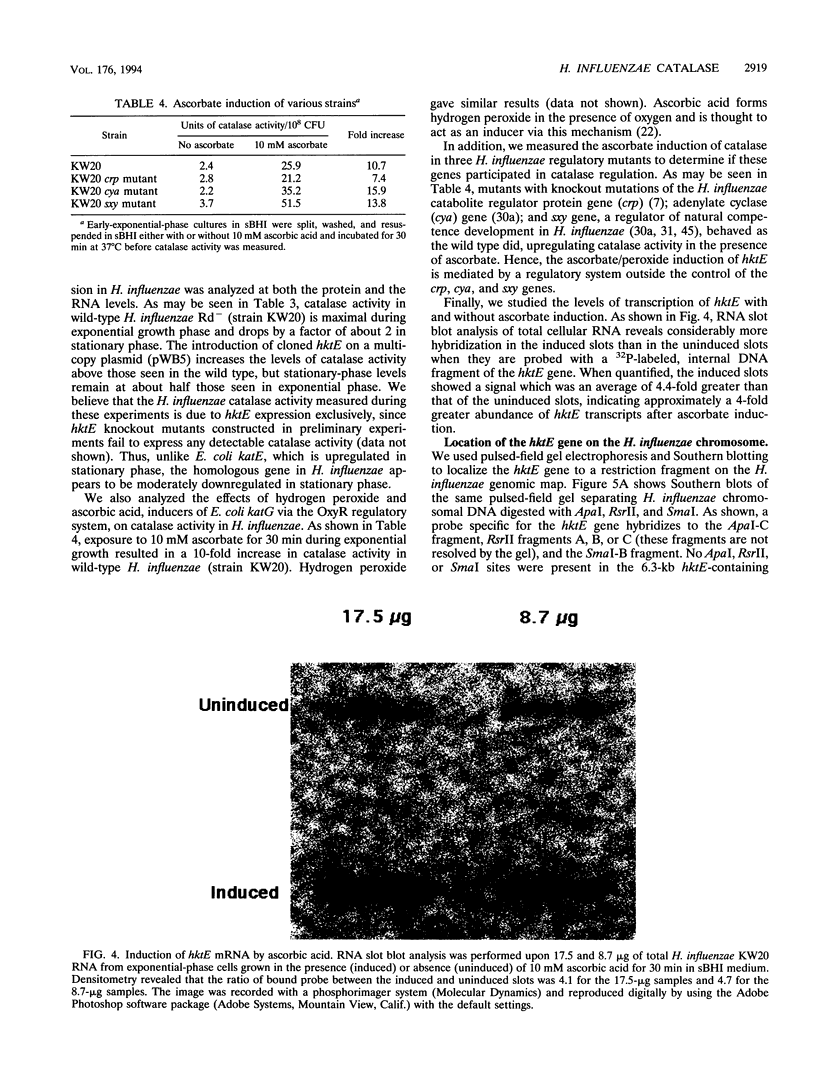
Bacterial catalases are induced by exposure to peroxide (e.g., Escherichia coli katG) or entry into stationary phase (e.g., E. coli katE). To study regulatory systems in Haemophilus influenzae, we complemented an E. coli rpoS mutant, which is unable to induce katE in stationary phase, with a plasmid library of H. influenzae Rd- chromosomal DNA. Nineteen complementing clones with a catalase-positive phenotype were obtained and characterized after screening about 10(5) transformants. All carried the same structural gene for an H. influenzae catalase. The DNA sequence of this gene, called hktE, encodes a 508-amino-acid polypeptide with strong homology to eukaryotic catalases and E. coli katE. However, hktE is regulated like E. coli katG, with catalase activity increasing 10-fold and hktE mRNA levels increasing 4-fold upon exposure to ascorbic acid, which serves to generate hydrogen peroxide. Mutations in the known global regulatory genes of H. influenzae--crp, cya, and sxy--do not affect the inducibility of hktE. The hktE gene maps to a 225-kb segment of the H. influenzae chromosome in a region encoding resistance to spectinomycin.
Full text
PDF



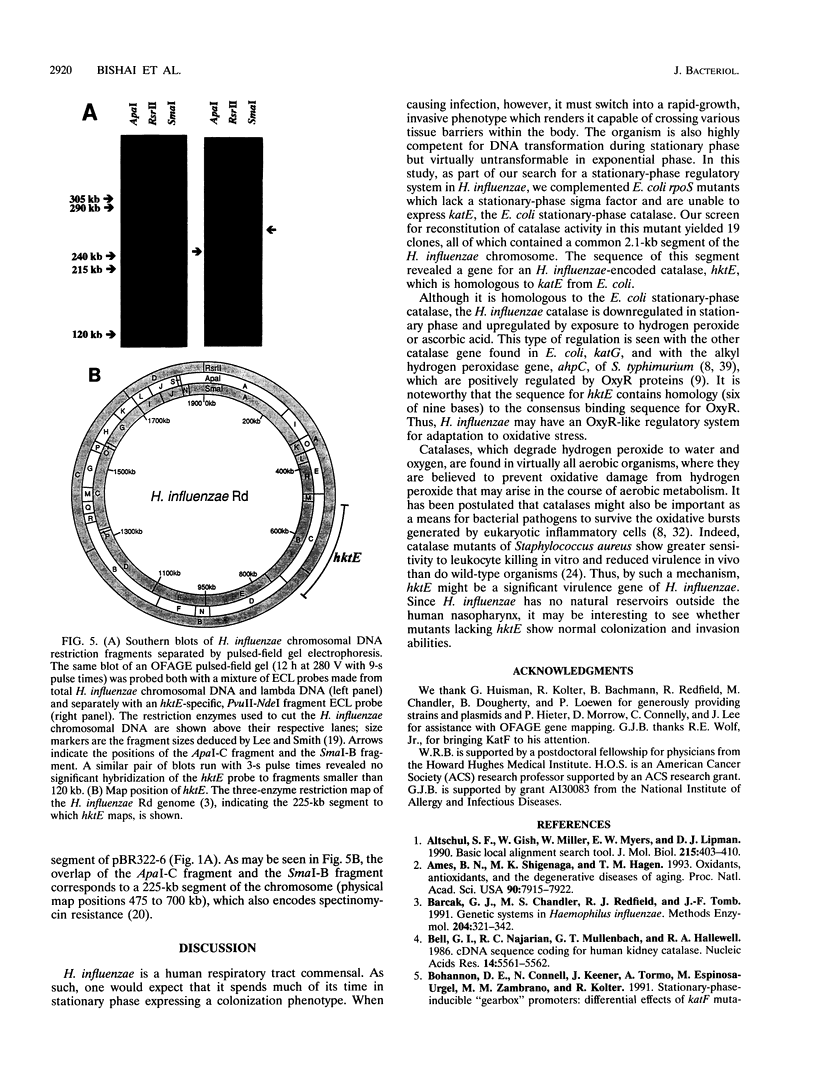

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. Basic local alignment search tool. J Mol Biol. 1990 Oct 5;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Ames B. N., Shigenaga M. K., Hagen T. M. Oxidants, antioxidants, and the degenerative diseases of aging. Proc Natl Acad Sci U S A. 1993 Sep 1;90(17):7915–7922. doi: 10.1073/pnas.90.17.7915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barcak G. J., Chandler M. S., Redfield R. J., Tomb J. F. Genetic systems in Haemophilus influenzae. Methods Enzymol. 1991;204:321–342. doi: 10.1016/0076-6879(91)04016-h. [DOI] [PubMed] [Google Scholar]
- Bell G. I., Najarian R. C., Mullenbach G. T., Hallewell R. A. cDNA sequence coding for human kidney catalase. Nucleic Acids Res. 1986 Jul 11;14(13):5561–5562. [PMC free article] [PubMed] [Google Scholar]
- Bol D. K., Yasbin R. E. The isolation, cloning and identification of a vegetative catalase gene from Bacillus subtilis. Gene. 1991 Dec 20;109(1):31–37. doi: 10.1016/0378-1119(91)90585-y. [DOI] [PubMed] [Google Scholar]
- Chandler M. S. The gene encoding cAMP receptor protein is required for competence development in Haemophilus influenzae Rd. Proc Natl Acad Sci U S A. 1992 Mar 1;89(5):1626–1630. doi: 10.1073/pnas.89.5.1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christman M. F., Morgan R. W., Jacobson F. S., Ames B. N. Positive control of a regulon for defenses against oxidative stress and some heat-shock proteins in Salmonella typhimurium. Cell. 1985 Jul;41(3):753–762. doi: 10.1016/s0092-8674(85)80056-8. [DOI] [PubMed] [Google Scholar]
- Christman M. F., Storz G., Ames B. N. OxyR, a positive regulator of hydrogen peroxide-inducible genes in Escherichia coli and Salmonella typhimurium, is homologous to a family of bacterial regulatory proteins. Proc Natl Acad Sci U S A. 1989 May;86(10):3484–3488. doi: 10.1073/pnas.86.10.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchward G., Belin D., Nagamine Y. A pSC101-derived plasmid which shows no sequence homology to other commonly used cloning vectors. Gene. 1984 Nov;31(1-3):165–171. doi: 10.1016/0378-1119(84)90207-5. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. "A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity". Addendum. Anal Biochem. 1984 Feb;137(1):266–267. doi: 10.1016/0003-2697(84)90381-6. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Fita I., Rossmann M. G. The NADPH binding site on beef liver catalase. Proc Natl Acad Sci U S A. 1985 Mar;82(6):1604–1608. doi: 10.1073/pnas.82.6.1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fita I., Rossmann M. G. The active center of catalase. J Mol Biol. 1985 Sep 5;185(1):21–37. doi: 10.1016/0022-2836(85)90180-9. [DOI] [PubMed] [Google Scholar]
- Hengge-Aronis R. Survival of hunger and stress: the role of rpoS in early stationary phase gene regulation in E. coli. Cell. 1993 Jan 29;72(2):165–168. doi: 10.1016/0092-8674(93)90655-a. [DOI] [PubMed] [Google Scholar]
- Herriott R. M., Meyer E. M., Vogt M. Defined nongrowth media for stage II development of competence in Haemophilus influenzae. J Bacteriol. 1970 Feb;101(2):517–524. doi: 10.1128/jb.101.2.517-524.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn M. E., Smith H. O. Transformation in Haemophilus: a problem in membrane biology. J Membr Biol. 1984;81(2):89–103. doi: 10.1007/BF01868974. [DOI] [PubMed] [Google Scholar]
- Lange R., Hengge-Aronis R. Identification of a central regulator of stationary-phase gene expression in Escherichia coli. Mol Microbiol. 1991 Jan;5(1):49–59. doi: 10.1111/j.1365-2958.1991.tb01825.x. [DOI] [PubMed] [Google Scholar]
- Lee J. J., Smith H. O., Redfield R. J. Organization of the Haemophilus influenzae Rd genome. J Bacteriol. 1989 Jun;171(6):3016–3024. doi: 10.1128/jb.171.6.3016-3024.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. J., Smith H. O. Sizing of the Haemophilus influenzae Rd genome by pulsed-field agarose gel electrophoresis. J Bacteriol. 1988 Sep;170(9):4402–4405. doi: 10.1128/jb.170.9.4402-4405.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewen P. C. Isolation of catalase-deficient Escherichia coli mutants and genetic mapping of katE, a locus that affects catalase activity. J Bacteriol. 1984 Feb;157(2):622–626. doi: 10.1128/jb.157.2.622-626.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewen P. C., Switala J., Triggs-Raine B. L. Catalases HPI and HPII in Escherichia coli are induced independently. Arch Biochem Biophys. 1985 Nov 15;243(1):144–149. doi: 10.1016/0003-9861(85)90782-9. [DOI] [PubMed] [Google Scholar]
- Loewen P. C., Triggs B. L. Genetic mapping of katF, a locus that with katE affects the synthesis of a second catalase species in Escherichia coli. J Bacteriol. 1984 Nov;160(2):668–675. doi: 10.1128/jb.160.2.668-675.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandell G. L. Catalase, superoxide dismutase, and virulence of Staphylococcus aureus. In vitro and in vivo studies with emphasis on staphylococcal--leukocyte interaction. J Clin Invest. 1975 Mar;55(3):561–566. doi: 10.1172/JCI107963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melik-Adamyan W. R., Barynin V. V., Vagin A. A., Borisov V. V., Vainshtein B. K., Fita I., Murthy M. R., Rossmann M. G. Comparison of beef liver and Penicillium vitale catalases. J Mol Biol. 1986 Mar 5;188(1):63–72. doi: 10.1016/0022-2836(86)90480-8. [DOI] [PubMed] [Google Scholar]
- Mulvey M. R., Loewen P. C. Nucleotide sequence of katF of Escherichia coli suggests KatF protein is a novel sigma transcription factor. Nucleic Acids Res. 1989 Dec 11;17(23):9979–9991. doi: 10.1093/nar/17.23.9979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov G. N., Melik-Adamyan W. R., Grebenko A. I., Barynin V. V., Vagin A. A., Vainshtein B. K., Dauter Z., Wilson K. S. Three-dimensional structure of catalase from Micrococcus lysodeikticus at 1.5 A resolution. FEBS Lett. 1992 Nov 9;312(2-3):127–131. doi: 10.1016/0014-5793(92)80919-8. [DOI] [PubMed] [Google Scholar]
- Murthy M. R., Reid T. J., 3rd, Sicignano A., Tanaka N., Rossmann M. G. Structure of beef liver catalase. J Mol Biol. 1981 Oct 25;152(2):465–499. doi: 10.1016/0022-2836(81)90254-0. [DOI] [PubMed] [Google Scholar]
- Redfield R. J. sxy-1, a Haemophilus influenzae mutation causing greatly enhanced spontaneous competence. J Bacteriol. 1991 Sep;173(18):5612–5618. doi: 10.1128/jb.173.18.5612-5618.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Root R. K., Cohen M. S. The microbicidal mechanisms of human neutrophils and eosinophils. Rev Infect Dis. 1981 May-Jun;3(3):565–598. doi: 10.1093/clinids/3.3.565. [DOI] [PubMed] [Google Scholar]
- Rorth M., Jensen P. K. Determination of catalase activity by means of the Clark oxygen electrode. Biochim Biophys Acta. 1967 May 16;139(1):171–173. doi: 10.1016/0005-2744(67)90124-6. [DOI] [PubMed] [Google Scholar]
- Roy P. H., Smith H. O. DNA methylases of Hemophilus influenzae Rd. I. Purification and properties. J Mol Biol. 1973 Dec 25;81(4):427–444. doi: 10.1016/0022-2836(73)90515-9. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder W. A., Shelton J. R., Shelton J. B., Robberson B., Apell G., Fang R. S., Bonaventura J. The complete amino acid sequence of bovine liver catalase and the partial sequence of bovine erythrocyte catalase. Arch Biochem Biophys. 1982 Mar;214(1):397–421. doi: 10.1016/0003-9861(82)90044-3. [DOI] [PubMed] [Google Scholar]
- Schuler G. D., Altschul S. F., Lipman D. J. A workbench for multiple alignment construction and analysis. Proteins. 1991;9(3):180–190. doi: 10.1002/prot.340090304. [DOI] [PubMed] [Google Scholar]
- Tartaglia L. A., Storz G., Ames B. N. Identification and molecular analysis of oxyR-regulated promoters important for the bacterial adaptation to oxidative stress. J Mol Biol. 1989 Dec 20;210(4):709–719. doi: 10.1016/0022-2836(89)90104-6. [DOI] [PubMed] [Google Scholar]
- Touati E., Dassa E., Boquet P. L. Pleiotropic mutations in appR reduce pH 2.5 acid phosphatase expression and restore succinate utilisation in CRP-deficient strains of Escherichia coli. Mol Gen Genet. 1986 Feb;202(2):257–264. doi: 10.1007/BF00331647. [DOI] [PubMed] [Google Scholar]
- Vainshtein B. K., Melik-Adamyan W. R., Barynin V. V., Vagin A. A., Grebenko A. I., Borisov V. V., Bartels K. S., Fita I., Rossmann M. G. Three-dimensional structure of catalase from Penicillium vitale at 2.0 A resolution. J Mol Biol. 1986 Mar 5;188(1):49–61. doi: 10.1016/0022-2836(86)90479-1. [DOI] [PubMed] [Google Scholar]
- Wilcox K. W., Smith H. O. Isolation and characterization of mutants of Haemophilus influenzae deficient in an adenosine 5'-triphosphate-dependent deoxyribonuclease activity. J Bacteriol. 1975 May;122(2):443–453. doi: 10.1128/jb.122.2.443-453.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Ossowski I., Hausner G., Loewen P. C. Molecular evolutionary analysis based on the amino acid sequence of catalase. J Mol Evol. 1993 Jul;37(1):71–76. doi: 10.1007/BF00170464. [DOI] [PubMed] [Google Scholar]
- von Ossowski I., Mulvey M. R., Leco P. A., Borys A., Loewen P. C. Nucleotide sequence of Escherichia coli katE, which encodes catalase HPII. J Bacteriol. 1991 Jan;173(2):514–520. doi: 10.1128/jb.173.2.514-520.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]