Abstract
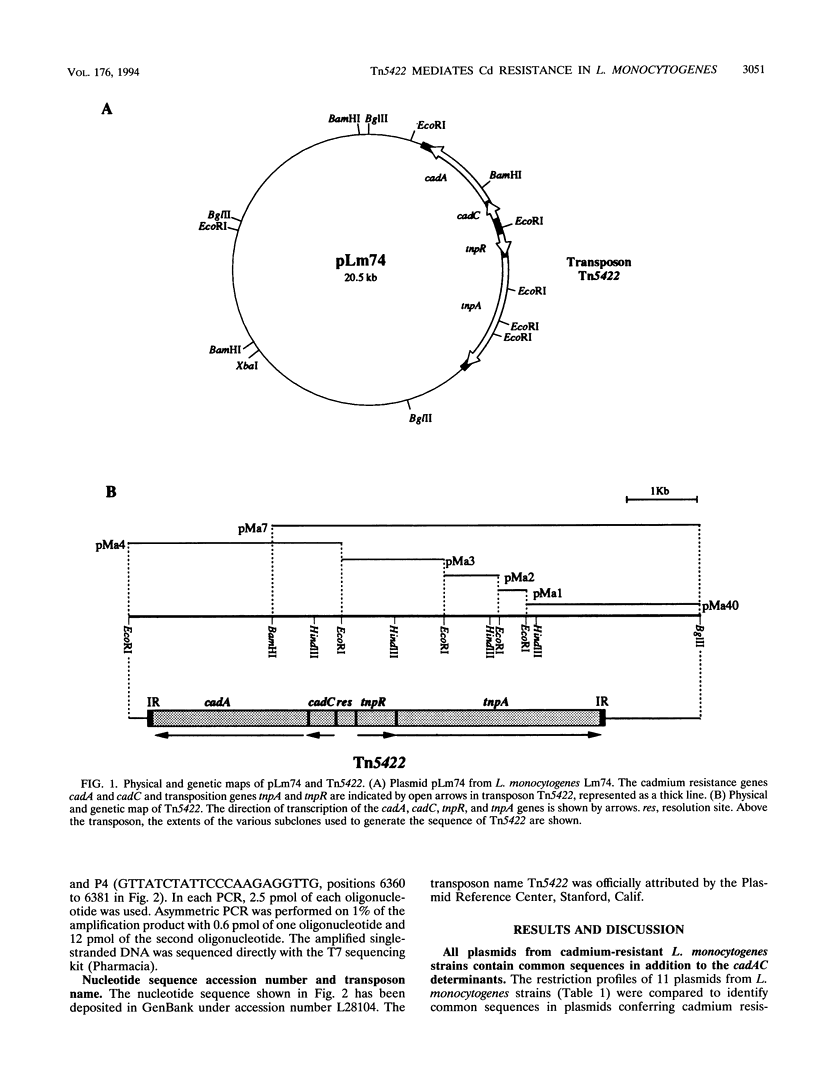
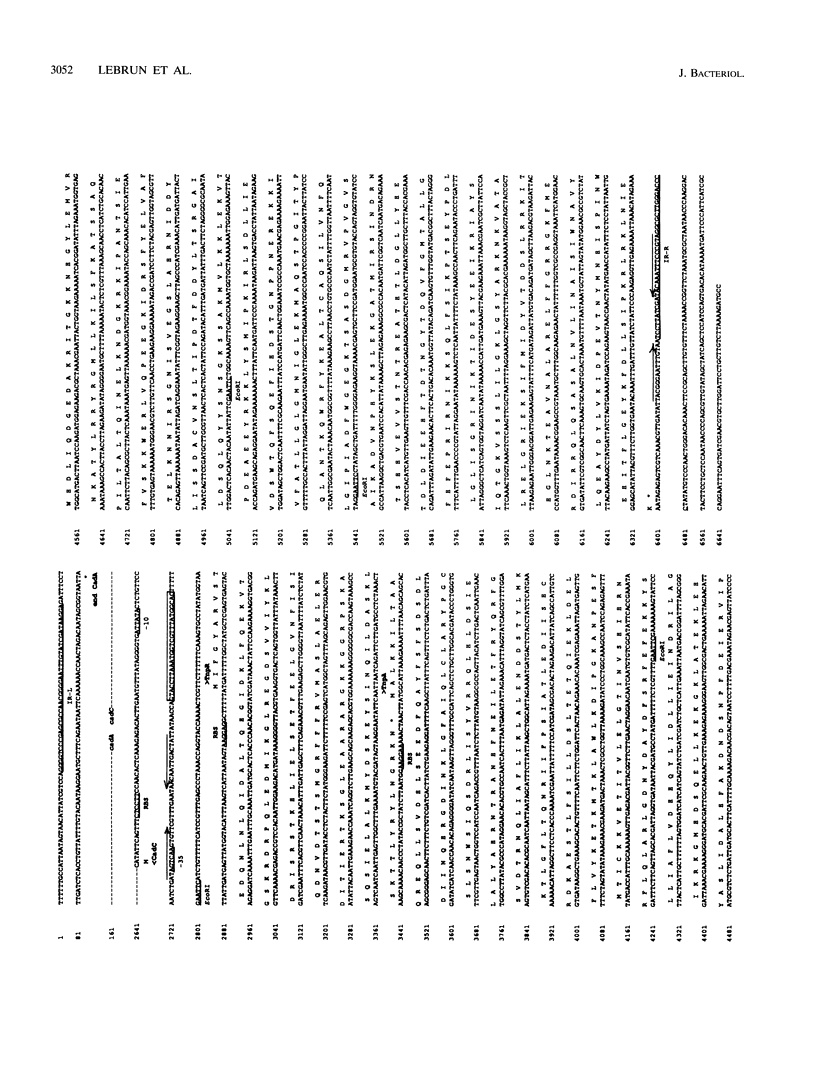
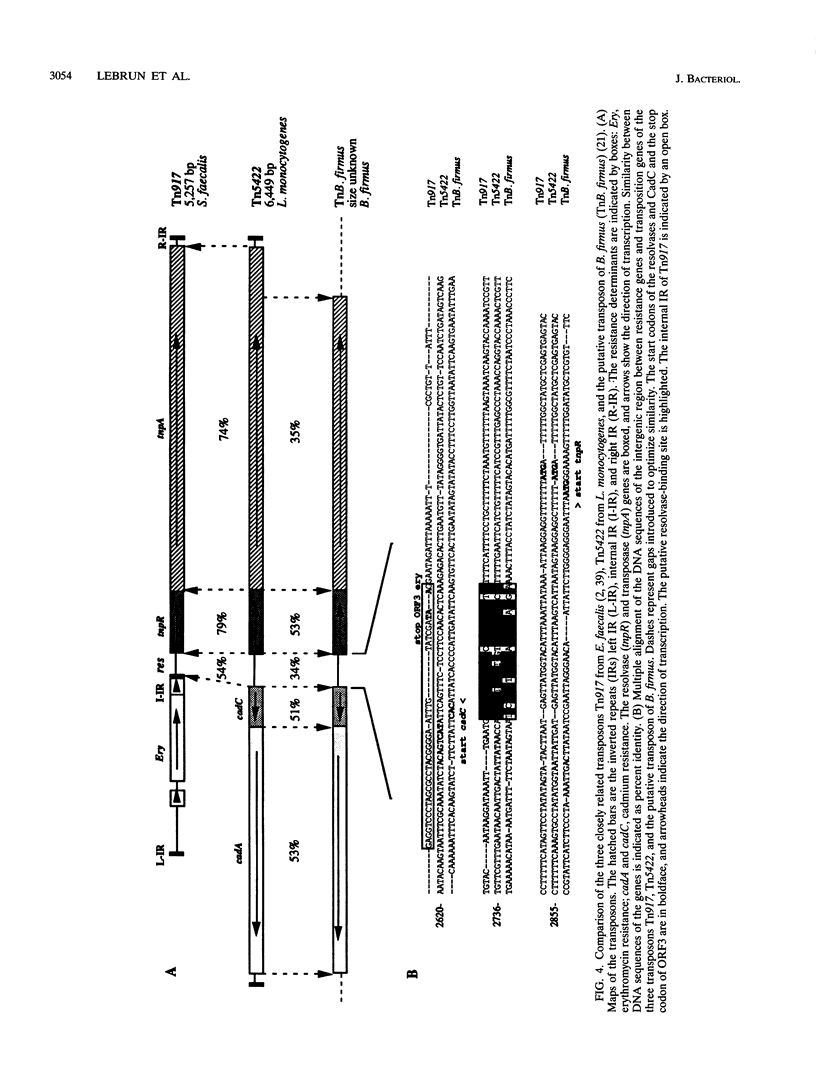
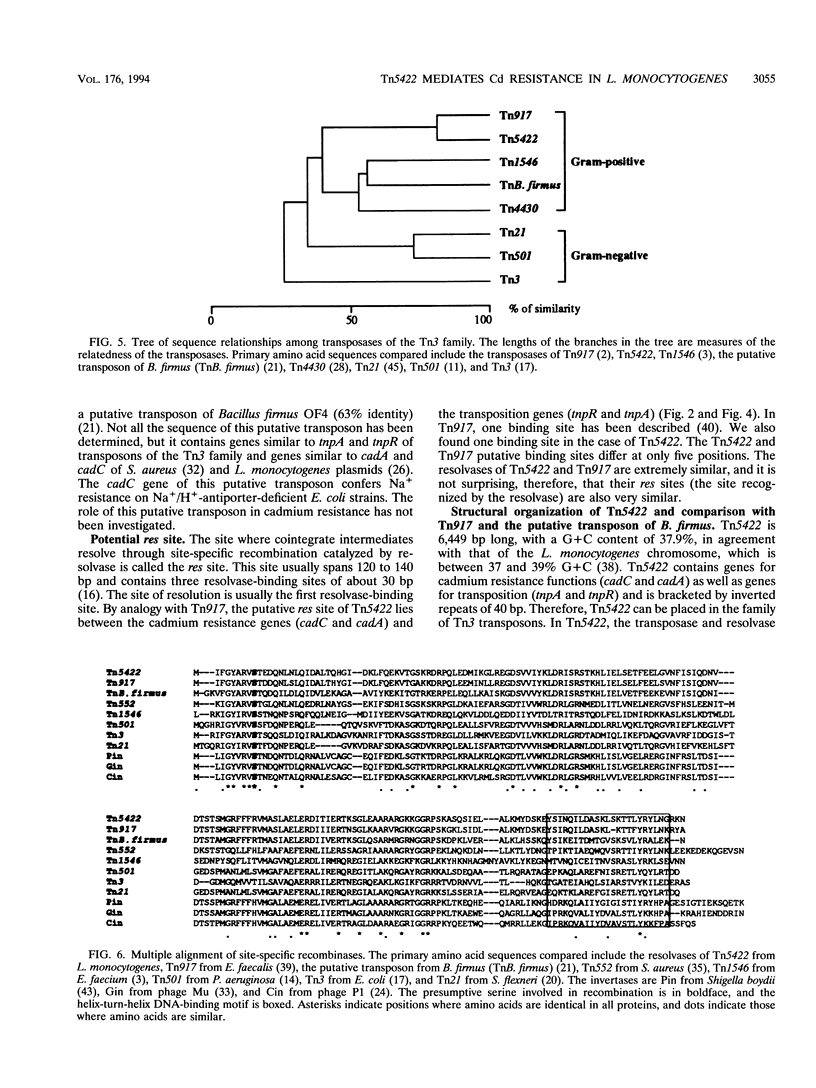
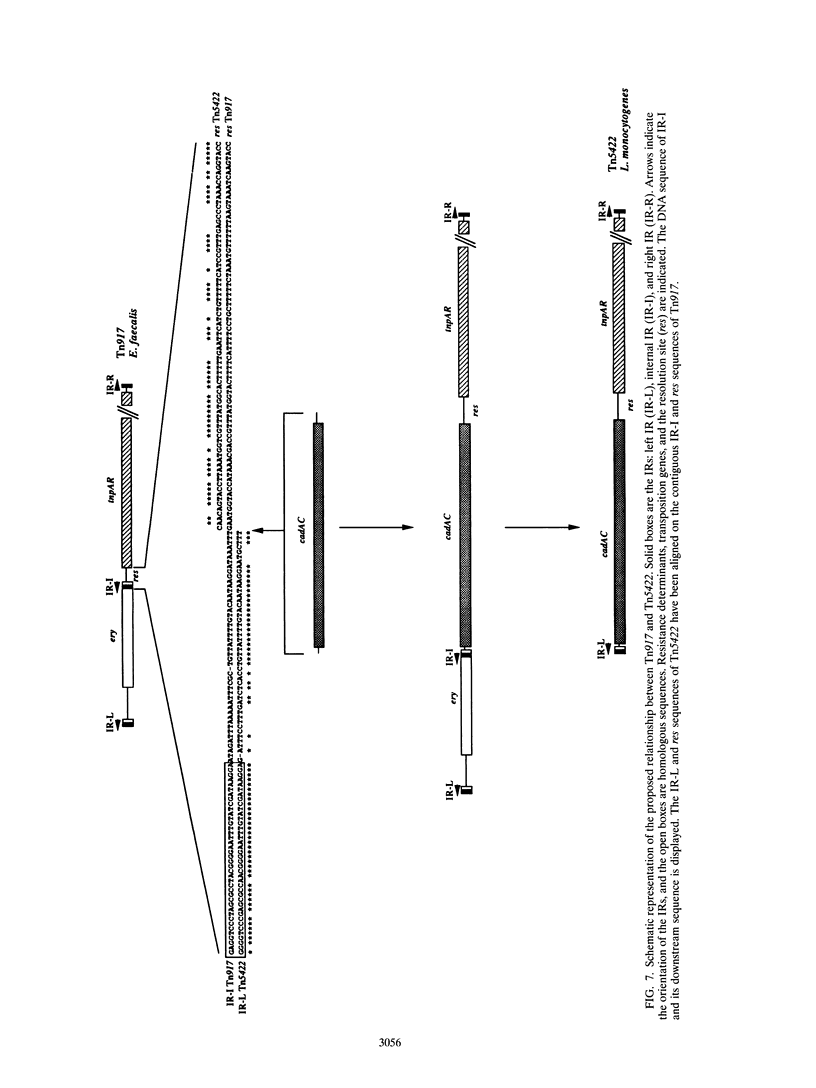
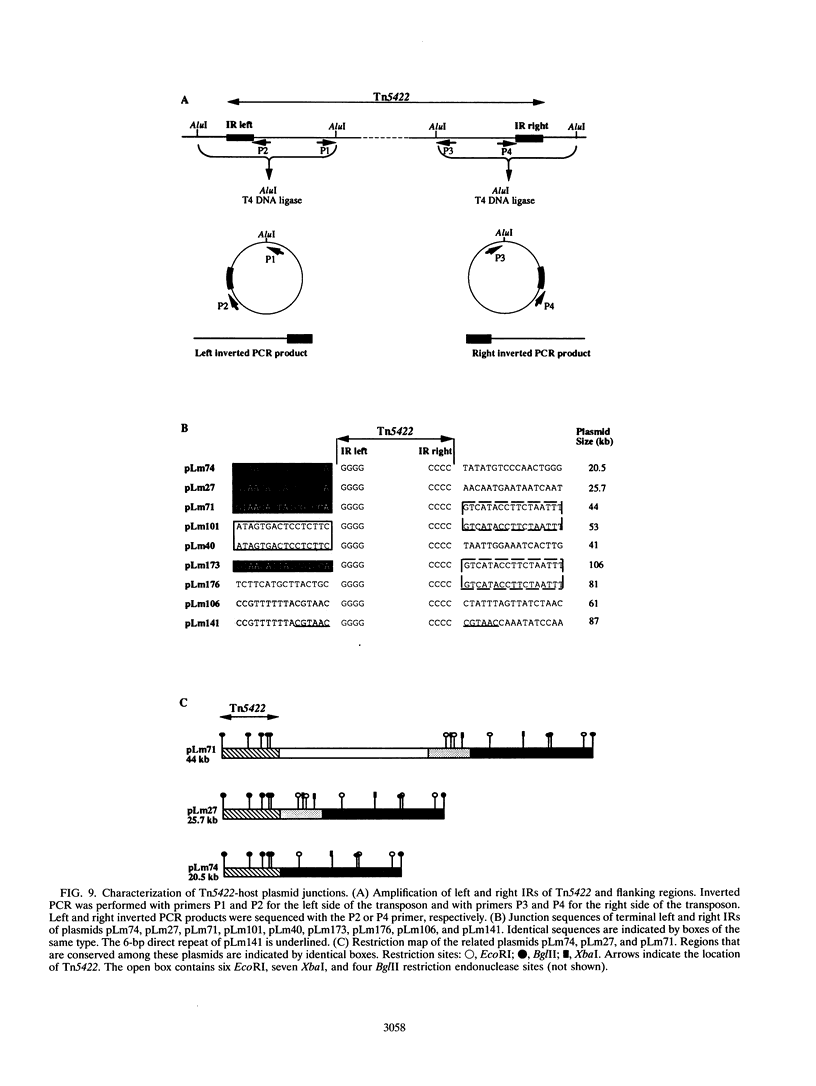
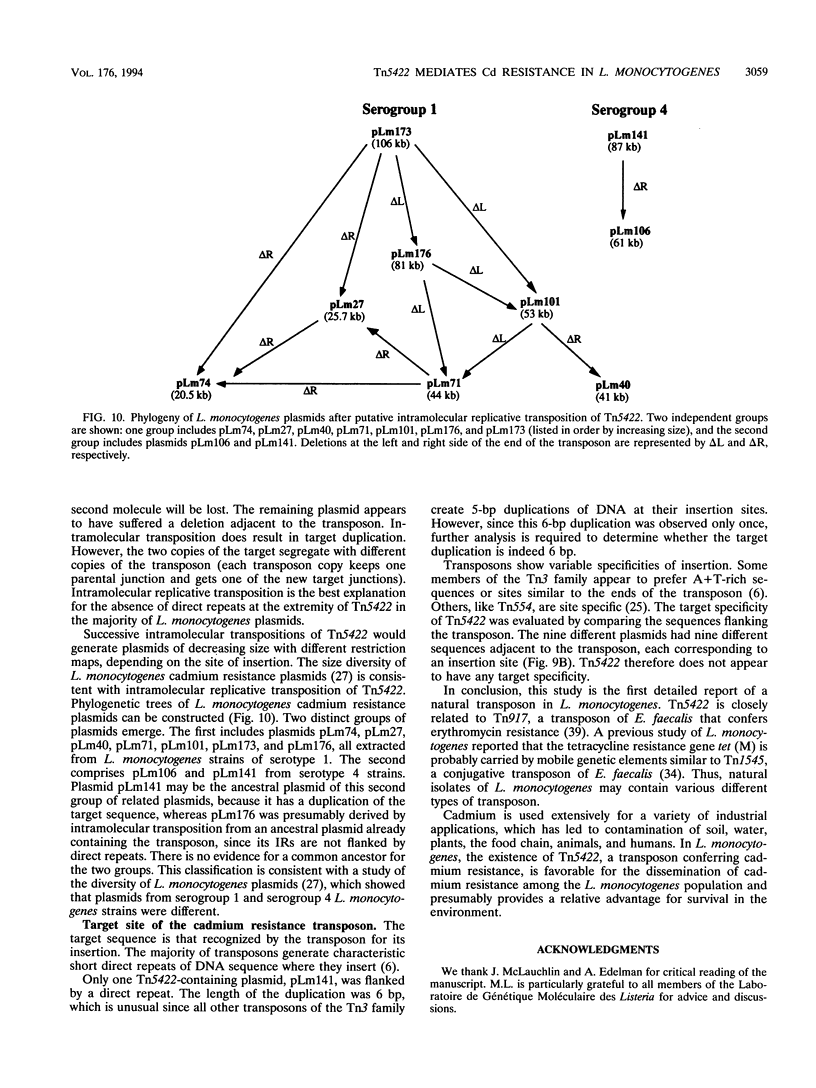
The complete (6,449-bp) nucleotide sequence of the first-described natural transposon of Listeria monocytogenes, designated Tn5422, was determined. Tn5422 is a transposon of the Tn3 family delineated by imperfect inverted repeats (IRs) of 40 bp. It contains two genes which confer cadmium resistance (M. Lebrun, A. Audurier, and P. Cossart, J. Bacteriol. 176:3040-3048, 1994) and two open reading frames that encode a transposase (TnpA) and a resolvase (TnpR) of 971 and 184 amino acids, respectively. The cadmium resistance genes and the transposition genes are transcribed in opposite directions and are separated by a putative recombination site (res). The structural elements presumed to be involved in transposition of Tn5422 (IRs, transposase, resolvase, and res) are very similar to those of Tn917, suggesting a common origin. The transposition genes were not induced by cadmium. Analysis of sequences surrounding Tn5422 in nine different plasmids of L. monocytogenes indicated that Tn5422 is a functional transposon, capable of intramolecular replicative transposition, generating deletions. This transposition process is probably the reason for the size diversity of the L. monocytogenes plasmids. Restriction analysis and Southern hybridization revealed the presence of Tn5422 in all the plasmid-mediated cadmium-resistant L. monocytogenes strains tested but not in strains encoding cadmium resistance on the chromosome.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. Basic local alignment search tool. J Mol Biol. 1990 Oct 5;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- An F. Y., Clewell D. B. Tn917 transposase. Sequence correction reveals a single open reading frame corresponding to the tnpA determinant of Tn3-family elements. Plasmid. 1991 Mar;25(2):121–124. doi: 10.1016/0147-619x(91)90023-p. [DOI] [PubMed] [Google Scholar]
- Arthur M., Molinas C., Depardieu F., Courvalin P. Characterization of Tn1546, a Tn3-related transposon conferring glycopeptide resistance by synthesis of depsipeptide peptidoglycan precursors in Enterococcus faecium BM4147. J Bacteriol. 1993 Jan;175(1):117–127. doi: 10.1128/jb.175.1.117-127.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett P. M., Grinsted J., Choi C. L., Richmond M. H. Characterisation of Tn501, a transposon determining resistance to mercuric ions. Mol Gen Genet. 1978 Feb 7;159(1):101–106. doi: 10.1007/BF00401753. [DOI] [PubMed] [Google Scholar]
- Bishop R., Sherratt D. Transposon Tn1 intra-molecular transposition. Mol Gen Genet. 1984;196(1):117–122. doi: 10.1007/BF00334102. [DOI] [PubMed] [Google Scholar]
- Borer P. N., Dengler B., Tinoco I., Jr, Uhlenbeck O. C. Stability of ribonucleic acid double-stranded helices. J Mol Biol. 1974 Jul 15;86(4):843–853. doi: 10.1016/0022-2836(74)90357-x. [DOI] [PubMed] [Google Scholar]
- Brown N. L., Choi C. L., Grinsted J., Richmond M. H., Whitehead P. R. Nucleotide sequences at the ends of the mercury resistance transposon, Tn501. Nucleic Acids Res. 1980 May 10;8(9):1933–1945. doi: 10.1093/nar/8.9.1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown N. L., Winnie J. N., Fritzinger D., Pridmore R. D. The nucleotide sequence of the tnpA gene completes the sequence of the Pseudomonas transposon Tn501. Nucleic Acids Res. 1985 Aug 12;13(15):5657–5669. doi: 10.1093/nar/13.15.5657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadaban M. J., Cohen S. N. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J Mol Biol. 1980 Apr;138(2):179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- Diver W. P., Grinsted J., Fritzinger D. C., Brown N. L., Altenbuchner J., Rogowsky P., Schmitt R. DNA sequences of and complementation by the tnpR genes of Tn21, Tn501 and Tn1721. Mol Gen Genet. 1983;191(2):189–193. doi: 10.1007/BF00334812. [DOI] [PubMed] [Google Scholar]
- Evans L. R., Brown N. L. Construction of hybrid Tn501/Tn21 transposases in vivo: identification of a region of transposase conferring specificity of recognition of the 38-bp terminal inverted repeats. EMBO J. 1987 Sep;6(9):2849–2853. doi: 10.1002/j.1460-2075.1987.tb02582.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grindley N. D., Lauth M. R., Wells R. G., Wityk R. J., Salvo J. J., Reed R. R. Transposon-mediated site-specific recombination: identification of three binding sites for resolvase at the res sites of gamma delta and Tn3. Cell. 1982 Aug;30(1):19–27. doi: 10.1016/0092-8674(82)90007-1. [DOI] [PubMed] [Google Scholar]
- Heffron F., McCarthy B. J., Ohtsubo H., Ohtsubo E. DNA sequence analysis of the transposon Tn3: three genes and three sites involved in transposition of Tn3. Cell. 1979 Dec;18(4):1153–1163. doi: 10.1016/0092-8674(79)90228-9. [DOI] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984 Jun;28(3):351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- Higgins D. G., Bleasby A. J., Fuchs R. CLUSTAL V: improved software for multiple sequence alignment. Comput Appl Biosci. 1992 Apr;8(2):189–191. doi: 10.1093/bioinformatics/8.2.189. [DOI] [PubMed] [Google Scholar]
- Hyde D. R., Tu C. P. tnpM: a novel regulatory gene that enhances Tn21 transposition and suppresses cointegrate resolution. Cell. 1985 Sep;42(2):629–638. doi: 10.1016/0092-8674(85)90120-5. [DOI] [PubMed] [Google Scholar]
- Ivey D. M., Guffanti A. A., Shen Z., Kudyan N., Krulwich T. A. The cadC gene product of alkaliphilic Bacillus firmus OF4 partially restores Na+ resistance to an Escherichia coli strain lacking an Na+/H+ antiporter (NhaA). J Bacteriol. 1992 Aug;174(15):4878–4884. doi: 10.1128/jb.174.15.4878-4884.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan S. A., Novick R. P. Terminal nucleotide sequences of Tn551, a transposon specifying erythromycin resistance in Staphylococcus aureus: homology with Tn3. Plasmid. 1980 Sep;4(2):148–154. doi: 10.1016/0147-619x(80)90004-9. [DOI] [PubMed] [Google Scholar]
- Kitts P., Symington L., Burke M., Reed R., Sherratt D. Transposon-specified site-specific recombination. Proc Natl Acad Sci U S A. 1982 Jan;79(1):46–50. doi: 10.1073/pnas.79.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klippel A., Cloppenborg K., Kahmann R. Isolation and characterization of unusual gin mutants. EMBO J. 1988 Dec 1;7(12):3983–3989. doi: 10.1002/j.1460-2075.1988.tb03286.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krolewski J. J., Murphy E., Novick R. P., Rush M. G. Site-specificity of the chromosomal insertion of Staphylococcus aureus transposon Tn554. J Mol Biol. 1981 Oct 15;152(1):19–33. doi: 10.1016/0022-2836(81)90093-0. [DOI] [PubMed] [Google Scholar]
- Lebrun M., Audurier A., Cossart P. Plasmid-borne cadmium resistance genes in Listeria monocytogenes are similar to cadA and cadC of Staphylococcus aureus and are induced by cadmium. J Bacteriol. 1994 May;176(10):3040–3048. doi: 10.1128/jb.176.10.3040-3048.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebrun M., Loulergue J., Chaslus-Dancla E., Audurier A. Plasmids in Listeria monocytogenes in relation to cadmium resistance. Appl Environ Microbiol. 1992 Sep;58(9):3183–3186. doi: 10.1128/aem.58.9.3183-3186.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahillon J., Lereclus D. Structural and functional analysis of Tn4430: identification of an integrase-like protein involved in the co-integrate-resolution process. EMBO J. 1988 May;7(5):1515–1526. doi: 10.1002/j.1460-2075.1988.tb02971.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mengaud J., Vicente M. F., Chenevert J., Pereira J. M., Geoffroy C., Gicquel-Sanzey B., Baquero F., Perez-Diaz J. C., Cossart P. Expression in Escherichia coli and sequence analysis of the listeriolysin O determinant of Listeria monocytogenes. Infect Immun. 1988 Apr;56(4):766–772. doi: 10.1128/iai.56.4.766-772.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nies D. H. Resistance to cadmium, cobalt, zinc, and nickel in microbes. Plasmid. 1992 Jan;27(1):17–28. doi: 10.1016/0147-619x(92)90003-s. [DOI] [PubMed] [Google Scholar]
- Nisen P. D., Kopecko D. J., Chou J., Cohen S. N. Site-specific DNA deletions occurring adjacent to the termini of a transposable ampicillin resistance element (Tn3). J Mol Biol. 1977 Dec 25;117(4):975–978. doi: 10.1016/s0022-2836(77)80008-9. [DOI] [PubMed] [Google Scholar]
- Nucifora G., Chu L., Misra T. K., Silver S. Cadmium resistance from Staphylococcus aureus plasmid pI258 cadA gene results from a cadmium-efflux ATPase. Proc Natl Acad Sci U S A. 1989 May;86(10):3544–3548. doi: 10.1073/pnas.86.10.3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plasterk R. H., Brinkman A., van de Putte P. DNA inversions in the chromosome of Escherichia coli and in bacteriophage Mu: relationship to other site-specific recombination systems. Proc Natl Acad Sci U S A. 1983 Sep;80(17):5355–5358. doi: 10.1073/pnas.80.17.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poyart-Salmeron C., Trieu-Cuot P., Carlier C., MacGowan A., McLauchlin J., Courvalin P. Genetic basis of tetracycline resistance in clinical isolates of Listeria monocytogenes. Antimicrob Agents Chemother. 1992 Feb;36(2):463–466. doi: 10.1128/aac.36.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland S. J., Dyke K. G. Characterization of the staphylococcal beta-lactamase transposon Tn552. EMBO J. 1989 Sep;8(9):2761–2773. doi: 10.1002/j.1460-2075.1989.tb08418.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw J. H., Clewell D. B. Complete nucleotide sequence of macrolide-lincosamide-streptogramin B-resistance transposon Tn917 in Streptococcus faecalis. J Bacteriol. 1985 Nov;164(2):782–796. doi: 10.1128/jb.164.2.782-796.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver S., Walderhaug M. Gene regulation of plasmid- and chromosome-determined inorganic ion transport in bacteria. Microbiol Rev. 1992 Mar;56(1):195–228. doi: 10.1128/mr.56.1.195-228.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomich P. K., An F. Y., Clewell D. B. Properties of erythromycin-inducible transposon Tn917 in Streptococcus faecalis. J Bacteriol. 1980 Mar;141(3):1366–1374. doi: 10.1128/jb.141.3.1366-1374.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tominaga A., Ikemizu S., Enomoto M. Site-specific recombinase genes in three Shigella subgroups and nucleotide sequences of a pinB gene and an invertible B segment from Shigella boydii. J Bacteriol. 1991 Jul;173(13):4079–4087. doi: 10.1128/jb.173.13.4079-4087.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tynecka Z., Gos Z., Zajac J. Energy-dependent efflux of cadmium coded by a plasmid resistance determinant in Staphylococcus aureus. J Bacteriol. 1981 Aug;147(2):313–319. doi: 10.1128/jb.147.2.313-319.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward E., Grinsted J. The nucleotide sequence of the tnpA gene of Tn21. Nucleic Acids Res. 1987 Feb 25;15(4):1799–1806. doi: 10.1093/nar/15.4.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- de la Cruz F., Grinsted J. Genetic and molecular characterization of Tn21, a multiple resistance transposon from R100.1. J Bacteriol. 1982 Jul;151(1):222–228. doi: 10.1128/jb.151.1.222-228.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]



