Abstract
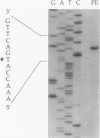
The restricted facultative methylotroph Methylophilus sp. strain DM11 utilizes dichloromethane as the sole carbon and energy source. It differs from other dichloromethane-utilizing methylotrophs by faster growth on this substrate and by possession of a group B dichloromethane dehalogenase catalyzing dechlorination at a fivefold-higher rate than the group A enzymes of slow-growing strains. We isolated dcmA, the structural gene of the strain DM11 dichloromethane dehalogenase, to elucidate its relationship to the previously characterized dcmA gene of Methylobacterium sp. strain DM4, which encodes a group A enzyme. Nucleotide sequence determination of dcmA from strain DM11 predicts a protein of 267 amino acids, corresponding to a molecular mass of 31,197 Da. The 5' terminus of in vivo dcmA transcripts was determined by primer extension to be 70 bp upstream of the translation initiation codon. It was preceded by a putative promoter sequence with high resemblance to the Escherichia coli sigma 70 consensus promoter sequence. dcmA and 130 bp of its upstream sequence were brought under control of the tac promoter and expressed in E. coli to approximately 20% of the total cellular protein by induction with isopropylthiogalactopyranoside (IPTG) and growth at 25 degrees C. Expression at 37 degrees C led to massive formation of inclusion bodies. Comparison of the strain DM11 and strain DM4 dichloromethane dehalogenase sequences revealed 59% identity at the DNA level and 56% identity at the protein level, thus indicating an ancient divergence of the two enzymes. Both dehalogenases are more closely related to eukaryotic class theta glutathione S-transferases than to a number of bacterial glutathione S-transferases.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arca P., Hardisson C., Suárez J. E. Purification of a glutathione S-transferase that mediates fosfomycin resistance in bacteria. Antimicrob Agents Chemother. 1990 May;34(5):844–848. doi: 10.1128/aac.34.5.844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blocki F. A., Ellis L. B., Wackett L. P. MIF protein are theta-class glutathione S-transferase homologs. Protein Sci. 1993 Dec;2(12):2095–2102. doi: 10.1002/pro.5560021210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Del Sal G., Manfioletti G., Schneider C. A one-tube plasmid DNA mini-preparation suitable for sequencing. Nucleic Acids Res. 1988 Oct 25;16(20):9878–9878. doi: 10.1093/nar/16.20.9878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Fürste J. P., Pansegrau W., Frank R., Blöcker H., Scholz P., Bagdasarian M., Lanka E. Molecular cloning of the plasmid RP4 primase region in a multi-host-range tacP expression vector. Gene. 1986;48(1):119–131. doi: 10.1016/0378-1119(86)90358-6. [DOI] [PubMed] [Google Scholar]
- Grove G., Zarlengo R. P., Timmerman K. P., Li N. Q., Tam M. F., Tu C. P. Characterization and heterospecific expression of cDNA clones of genes in the maize GSH S-transferase multigene family. Nucleic Acids Res. 1988 Jan 25;16(2):425–438. doi: 10.1093/nar/16.2.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983 Jun 5;166(4):557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Harley C. B., Reynolds R. P. Analysis of E. coli promoter sequences. Nucleic Acids Res. 1987 Mar 11;15(5):2343–2361. doi: 10.1093/nar/15.5.2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji X., Zhang P., Armstrong R. N., Gilliland G. L. The three-dimensional structure of a glutathione S-transferase from the mu gene class. Structural analysis of the binary complex of isoenzyme 3-3 and glutathione at 2.2-A resolution. Biochemistry. 1992 Oct 27;31(42):10169–10184. doi: 10.1021/bi00157a004. [DOI] [PubMed] [Google Scholar]
- Kohler-Staub D., Leisinger T. Dichloromethane dehalogenase of Hyphomicrobium sp. strain DM2. J Bacteriol. 1985 May;162(2):676–681. doi: 10.1128/jb.162.2.676-681.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong K. H., Inoue H., Takahashi K. Non-essentiality of cysteine and histidine residues for the activity of human class PI glutathione S-transferase. Biochem Biophys Res Commun. 1991 Dec 16;181(2):748–755. doi: 10.1016/0006-291x(91)91254-a. [DOI] [PubMed] [Google Scholar]
- La Roche S. D., Leisinger T. Identification of dcmR, the regulatory gene governing expression of dichloromethane dehalogenase in Methylobacterium sp. strain DM4. J Bacteriol. 1991 Nov;173(21):6714–6721. doi: 10.1128/jb.173.21.6714-6721.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Roche S. D., Leisinger T. Sequence analysis and expression of the bacterial dichloromethane dehalogenase structural gene, a member of the glutathione S-transferase supergene family. J Bacteriol. 1990 Jan;172(1):164–171. doi: 10.1128/jb.172.1.164-171.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lai H. C., Grove G., Tu C. P. Cloning and sequence analysis of a cDNA for a rat liver glutathione S-transferase Yb subunit. Nucleic Acids Res. 1986 Aug 11;14(15):6101–6114. doi: 10.1093/nar/14.15.6101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leaver M. J., Scott K., George S. G. Cloning and characterization of the major hepatic glutathione S-transferase from a marine teleost flatfish, the plaice (Pleuronectes platessa), with structural similarities to plant, insect and mammalian Theta class isoenzymes. Biochem J. 1993 May 15;292(Pt 1):189–195. doi: 10.1042/bj2920189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lidstrom M. E., Stirling D. I. Methylotrophs: genetics and commercial applications. Annu Rev Microbiol. 1990;44:27–58. doi: 10.1146/annurev.mi.44.100190.000331. [DOI] [PubMed] [Google Scholar]
- Liu S., Zhang P., Ji X., Johnson W. W., Gilliland G. L., Armstrong R. N. Contribution of tyrosine 6 to the catalytic mechanism of isoenzyme 3-3 of glutathione S-transferase. J Biol Chem. 1992 Mar 5;267(7):4296–4299. [PubMed] [Google Scholar]
- Mannervik B., Awasthi Y. C., Board P. G., Hayes J. D., Di Ilio C., Ketterer B., Listowsky I., Morgenstern R., Muramatsu M., Pearson W. R. Nomenclature for human glutathione transferases. Biochem J. 1992 Feb 15;282(Pt 1):305–306. doi: 10.1042/bj2820305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masai E., Katayama Y., Kubota S., Kawai S., Yamasaki M., Morohoshi N. A bacterial enzyme degrading the model lignin compound beta-etherase is a member of the glutathione-S-transferase superfamily. FEBS Lett. 1993 May 24;323(1-2):135–140. doi: 10.1016/0014-5793(93)81465-c. [DOI] [PubMed] [Google Scholar]
- Meyer D. J., Coles B., Pemble S. E., Gilmore K. S., Fraser G. M., Ketterer B. Theta, a new class of glutathione transferases purified from rat and man. Biochem J. 1991 Mar 1;274(Pt 2):409–414. doi: 10.1042/bj2740409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mignogna G., Allocati N., Aceto A., Piccolomini R., Di Ilio C., Barra D., Martini F. The amino acid sequence of glutathione transferase from Proteus mirabilis, a prototype of a new class of enzymes. Eur J Biochem. 1993 Feb 1;211(3):421–425. doi: 10.1111/j.1432-1033.1993.tb17566.x. [DOI] [PubMed] [Google Scholar]
- Ogura K., Nishiyama T., Okada T., Kajital J., Narihata H., Watabe T., Hiratsuka A., Watabe T. Molecular cloning and amino acid sequencing of rat liver class theta glutathione S-transferase Yrs-Yrs inactivating reactive sulfate esters of carcinogenic arylmethanols. Biochem Biophys Res Commun. 1991 Dec 31;181(3):1294–1300. doi: 10.1016/0006-291x(91)92079-y. [DOI] [PubMed] [Google Scholar]
- Orser C. S., Dutton J., Lange C., Jablonski P., Xun L., Hargis M. Characterization of a Flavobacterium glutathione S-transferase gene involved reductive dechlorination. J Bacteriol. 1993 May;175(9):2640–2644. doi: 10.1128/jb.175.9.2640-2644.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pemble S. E., Taylor J. B. An evolutionary perspective on glutathione transferases inferred from class-theta glutathione transferase cDNA sequences. Biochem J. 1992 Nov 1;287(Pt 3):957–963. doi: 10.1042/bj2870957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinemer P., Dirr H. W., Ladenstein R., Huber R., Lo Bello M., Federici G., Parker M. W. Three-dimensional structure of class pi glutathione S-transferase from human placenta in complex with S-hexylglutathione at 2.8 A resolution. J Mol Biol. 1992 Sep 5;227(1):214–226. doi: 10.1016/0022-2836(92)90692-d. [DOI] [PubMed] [Google Scholar]
- Reinemer P., Dirr H. W., Ladenstein R., Schäffer J., Gallay O., Huber R. The three-dimensional structure of class pi glutathione S-transferase in complex with glutathione sulfonate at 2.3 A resolution. EMBO J. 1991 Aug;10(8):1997–2005. doi: 10.1002/j.1460-2075.1991.tb07729.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholtz R., Wackett L. P., Egli C., Cook A. M., Leisinger T. Dichloromethane dehalogenase with improved catalytic activity isolated from a fast-growing dichloromethane-utilizing bacterium. J Bacteriol. 1988 Dec;170(12):5698–5704. doi: 10.1128/jb.170.12.5698-5704.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Short J. M., Fernandez J. M., Sorge J. A., Huse W. D. Lambda ZAP: a bacteriophage lambda expression vector with in vivo excision properties. Nucleic Acids Res. 1988 Aug 11;16(15):7583–7600. doi: 10.1093/nar/16.15.7583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinning I., Kleywegt G. J., Cowan S. W., Reinemer P., Dirr H. W., Huber R., Gilliland G. L., Armstrong R. N., Ji X., Board P. G. Structure determination and refinement of human alpha class glutathione transferase A1-1, and a comparison with the Mu and Pi class enzymes. J Mol Biol. 1993 Jul 5;232(1):192–212. doi: 10.1006/jmbi.1993.1376. [DOI] [PubMed] [Google Scholar]
- Stenberg G., Björnestedt R., Mannervik B. Heterologous expression of recombinant human glutathione transferase A1-1 from a hepatoma cell line. Protein Expr Purif. 1992 Feb;3(1):80–84. doi: 10.1016/1046-5928(92)90060-a. [DOI] [PubMed] [Google Scholar]
- Takahashi Y., Nagata T. parB: an auxin-regulated gene encoding glutathione S-transferase. Proc Natl Acad Sci U S A. 1992 Jan 1;89(1):56–59. doi: 10.1073/pnas.89.1.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telakowski-Hopkins C. A., Rodkey J. A., Bennett C. D., Lu A. Y., Pickett C. B. Rat liver glutathione S-transferases. Construction of a cDNA clone complementary to a Yc mRNA and prediction of the complete amino acid sequence of a Yc subunit. J Biol Chem. 1985 May 10;260(9):5820–5825. [PubMed] [Google Scholar]
- Zhang P., Liu S., Shan S. O., Ji X., Gilliland G. L., Armstrong R. N. Modular mutagenesis of exons 1, 2, and 8 of a glutathione S-transferase from the mu class. Mechanistic and structural consequences for chimeras of isoenzyme 3-3. Biochemistry. 1992 Oct 27;31(42):10185–10193. doi: 10.1021/bi00157a005. [DOI] [PubMed] [Google Scholar]