Abstract
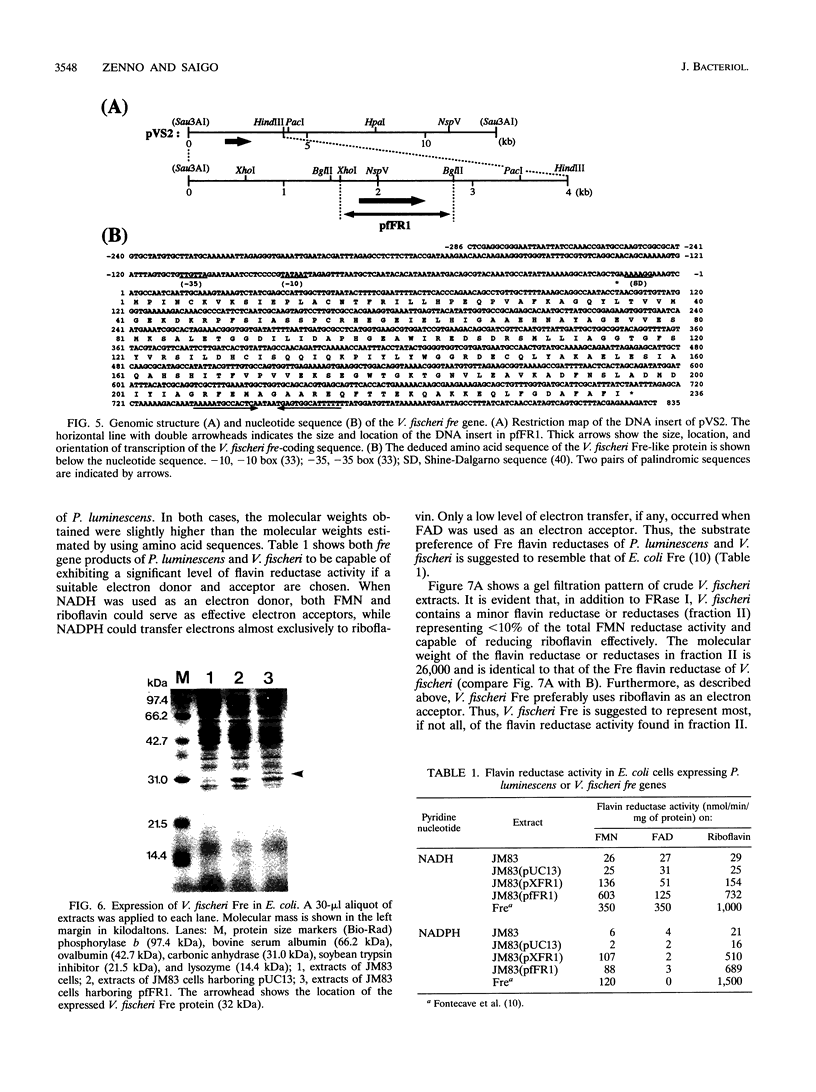
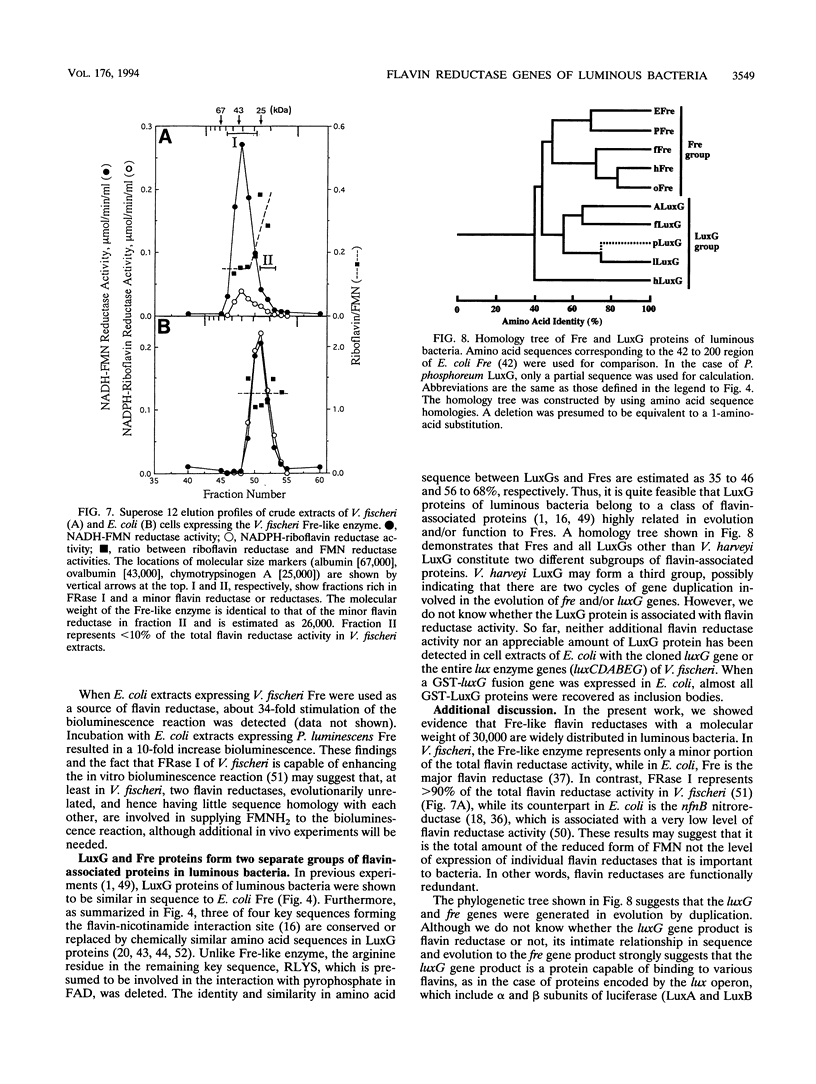
Genes encoding NAD(P)H-flavin oxidoreductases (flavin reductases) similar in both size and sequence to Fre, the most abundant flavin reductase in Escherichia coli, were identified in four species of luminous bacteria, Photorhabdus luminescens (ATCC 29999), Vibrio fischeri (ATCC 7744), Vibrio harveyi (ATCC 33843), and Vibrio orientalis (ATCC 33934). Nucleotide sequence analysis showed Fre-like flavin reductases in P. luminescens and V. fischeri to consist of 233 and 236 amino acids, respectively. As in E. coli Fre, Fre-like enzymes in luminous bacteria preferably used riboflavin as an electron acceptor when NADPH was used as an electron donor. These enzymes also were good suppliers of reduced flavin mononucleotide (FMNH2) to the bioluminescence reaction. In V. fischeri, the Fre-like enzyme is a minor flavin reductase representing < 10% of the total FMN reductase. That the V. fischeri Fre-like enzyme has no appreciable homology in amino acid sequence to the major flavin reductase in V. fischeri, FRase I, indicates that at least two different types of flavin reductases supply FMNH2 to the luminescence system in V. fischeri. Although Fre-like flavin reductases are highly similar in sequence to luxG gene products (LuxGs), Fre-like flavin reductases and LuxGs appear to constitute two separate groups of flavin-associated proteins.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andrews S. C., Shipley D., Keen J. N., Findlay J. B., Harrison P. M., Guest J. R. The haemoglobin-like protein (HMP) of Escherichia coli has ferrisiderophore reductase activity and its C-terminal domain shares homology with ferredoxin NADP+ reductases. FEBS Lett. 1992 May 18;302(3):247–252. doi: 10.1016/0014-5793(92)80452-m. [DOI] [PubMed] [Google Scholar]
- Anlezark G. M., Melton R. G., Sherwood R. F., Coles B., Friedlos F., Knox R. J. The bioactivation of 5-(aziridin-1-yl)-2,4-dinitrobenzamide (CB1954)--I. Purification and properties of a nitroreductase enzyme from Escherichia coli--a potential enzyme for antibody-directed enzyme prodrug therapy (ADEPT). Biochem Pharmacol. 1992 Dec 15;44(12):2289–2295. doi: 10.1016/0006-2952(92)90671-5. [DOI] [PubMed] [Google Scholar]
- Appleyard R K. Segregation of Lambda Lysogenicity during Bacterial Recombination in Escherichia Coli K12. Genetics. 1954 Jul;39(4):429–439. doi: 10.1093/genetics/39.4.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin T. O., Devine J. H., Heckel R. C., Lin J. W., Shadel G. S. The complete nucleotide sequence of the lux regulon of Vibrio fischeri and the luxABN region of Photobacterium leiognathi and the mechanism of control of bacterial bioluminescence. J Biolumin Chemilumin. 1989 Jul;4(1):326–341. doi: 10.1002/bio.1170040145. [DOI] [PubMed] [Google Scholar]
- Baldwin T. O., Treat M. L., Daubner S. C. Cloning and expression of the luxY gene from Vibrio fischeri strain Y-1 in Escherichia coli and complete amino acid sequence of the yellow fluorescent protein. Biochemistry. 1990 Jun 12;29(23):5509–5515. doi: 10.1021/bi00475a014. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Daubner S. C., Astorga A. M., Leisman G. B., Baldwin T. O. Yellow light emission of Vibrio fischeri strain Y-1: purification and characterization of the energy-accepting yellow fluorescent protein. Proc Natl Acad Sci U S A. 1987 Dec;84(24):8912–8916. doi: 10.1073/pnas.84.24.8912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duane W., Hastings J. W. Flavin mononucleotide reductase of luminous bacteria. Mol Cell Biochem. 1975 Jan 31;6(1):53–64. doi: 10.1007/BF01731866. [DOI] [PubMed] [Google Scholar]
- Fontecave M., Eliasson R., Reichard P. NAD(P)H:flavin oxidoreductase of Escherichia coli. A ferric iron reductase participating in the generation of the free radical of ribonucleotide reductase. J Biol Chem. 1987 Sep 5;262(25):12325–12331. [PubMed] [Google Scholar]
- Franklund C. V., Baron S. F., Hylemon P. B. Characterization of the baiH gene encoding a bile acid-inducible NADH:flavin oxidoreductase from Eubacterium sp. strain VPI 12708. J Bacteriol. 1993 May;175(10):3002–3012. doi: 10.1128/jb.175.10.3002-3012.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings J. W., Potrikus C. J., Gupta S. C., Kurfürst M., Makemson J. C. Biochemistry and physiology of bioluminescent bacteria. Adv Microb Physiol. 1985;26:235–291. doi: 10.1016/s0065-2911(08)60398-7. [DOI] [PubMed] [Google Scholar]
- Hattori M., Sakaki Y. Dideoxy sequencing method using denatured plasmid templates. Anal Biochem. 1986 Feb 1;152(2):232–238. doi: 10.1016/0003-2697(86)90403-3. [DOI] [PubMed] [Google Scholar]
- Illarionov B. A., Protopopova M. V., Karginov V. A., Mertvetsov N. P., Gitelson J. I. Nucleotide sequence of part of Photobacterium leiognathi lux region. Nucleic Acids Res. 1988 Oct 25;16(20):9855–9855. doi: 10.1093/nar/16.20.9855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jablonski E., DeLuca M. Purification and properties of the NADH and NADPH specific FMN oxidoreductases from Beneckea harveyi. Biochemistry. 1977 Jun 28;16(13):2932–2936. doi: 10.1021/bi00632a020. [DOI] [PubMed] [Google Scholar]
- Karplus P. A., Daniels M. J., Herriott J. R. Atomic structure of ferredoxin-NADP+ reductase: prototype for a structurally novel flavoenzyme family. Science. 1991 Jan 4;251(4989):60–66. [PubMed] [Google Scholar]
- Kita A., Kasai N., Kasai S., Nakaya T., Miki K. Crystallization and preliminary X-ray diffraction studies of a flavoprotein, FP390, from a luminescent bacterium, Photobacterium phosphoreum. J Biochem. 1991 Nov;110(5):748–750. doi: 10.1093/oxfordjournals.jbchem.a123652. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lee C. Y., Szittner R. B., Meighen E. A. The lux genes of the luminous bacterial symbiont, Photobacterium leiognathi, of the ponyfish. Nucleotide sequence, difference in gene organization, and high expression in mutant Escherichia coli. Eur J Biochem. 1991 Oct 1;201(1):161–167. doi: 10.1111/j.1432-1033.1991.tb16269.x. [DOI] [PubMed] [Google Scholar]
- Macheroux P., Schmidt K. U., Steinerstauch P., Ghisla S., Colepicolo P., Buntic R., Hastings J. W. Purification of the yellow fluorescent protein from Vibrio fischeri and identity of the flavin chromophore. Biochem Biophys Res Commun. 1987 Jul 15;146(1):101–106. doi: 10.1016/0006-291x(87)90696-6. [DOI] [PubMed] [Google Scholar]
- Maki M., Takano E., Mori H., Sato A., Murachi T., Hatanaka M. All four internally repetitive domains of pig calpastatin possess inhibitory activities against calpains I and II. FEBS Lett. 1987 Oct 19;223(1):174–180. doi: 10.1016/0014-5793(87)80531-8. [DOI] [PubMed] [Google Scholar]
- Mancini J. A., Boylan M., Soly R. R., Graham A. F., Meighen E. A. Cloning and expression of the Photobacterium phosphoreum luminescence system demonstrates a unique lux gene organization. J Biol Chem. 1988 Oct 5;263(28):14308–14314. [PubMed] [Google Scholar]
- Meighen E. A. Molecular biology of bacterial bioluminescence. Microbiol Rev. 1991 Mar;55(1):123–142. doi: 10.1128/mr.55.1.123-142.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Messing J., Vieira J. A new pair of M13 vectors for selecting either DNA strand of double-digest restriction fragments. Gene. 1982 Oct;19(3):269–276. doi: 10.1016/0378-1119(82)90016-6. [DOI] [PubMed] [Google Scholar]
- Moore S. A., James M. N., O'Kane D. J., Lee J. Crystal structure of a flavoprotein related to the subunits of bacterial luciferase. EMBO J. 1993 May;12(5):1767–1774. doi: 10.1002/j.1460-2075.1993.tb05824.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Kane D. J., Prasher D. C. Evolutionary origins of bacterial bioluminescence. Mol Microbiol. 1992 Feb;6(4):443–449. doi: 10.1111/j.1365-2958.1992.tb01488.x. [DOI] [PubMed] [Google Scholar]
- Puget K., Michelson A. M. Studies in bioluminescence. VII. Bacterial NADH: flavin mononucleotide oxidoreductase. Biochimie. 1972;54(9):1197–1204. doi: 10.1016/s0300-9084(72)80024-5. [DOI] [PubMed] [Google Scholar]
- Raibekas A. A. Green flavoprotein from P. leiognathi: purification, characterization and identification as the product of the lux G(N) gene. J Biolumin Chemilumin. 1991 Jul-Sep;6(3):169–176. doi: 10.1002/bio.1170060306. [DOI] [PubMed] [Google Scholar]
- Raleigh E. A., Wilson G. Escherichia coli K-12 restricts DNA containing 5-methylcytosine. Proc Natl Acad Sci U S A. 1986 Dec;83(23):9070–9074. doi: 10.1073/pnas.83.23.9070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg M., Court D. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu Rev Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- Saiki R. K., Gelfand D. H., Stoffel S., Scharf S. J., Higuchi R., Horn G. T., Mullis K. B., Erlich H. A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988 Jan 29;239(4839):487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- Sastry S. S., Jayaraman R. Nitrofurantoin-resistant mutants of Escherichia coli: isolation and mapping. Mol Gen Genet. 1984;196(2):379–380. doi: 10.1007/BF00328076. [DOI] [PubMed] [Google Scholar]
- Schmidt T. M., Kopecky K., Nealson K. H. Bioluminescence of the insect pathogen Xenorhabdus luminescens. Appl Environ Microbiol. 1989 Oct;55(10):2607–2612. doi: 10.1128/aem.55.10.2607-2612.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. The 3'-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soly R. R., Mancini J. A., Ferri S. R., Boylan M., Meighen E. A. A new lux gene in bioluminescent bacteria codes for a protein homologous to the bacterial luciferase subunits. Biochem Biophys Res Commun. 1988 Aug 30;155(1):351–358. doi: 10.1016/s0006-291x(88)81092-1. [DOI] [PubMed] [Google Scholar]
- Spyrou G., Haggård-Ljungquist E., Krook M., Jörnvall H., Nilsson E., Reichard P. Characterization of the flavin reductase gene (fre) of Escherichia coli and construction of a plasmid for overproduction of the enzyme. J Bacteriol. 1991 Jun;173(12):3673–3679. doi: 10.1128/jb.173.12.3673-3679.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swartzman A., Kapoor S., Graham A. F., Meighen E. A. A new Vibrio fischeri lux gene precedes a bidirectional termination site for the lux operon. J Bacteriol. 1990 Dec;172(12):6797–6802. doi: 10.1128/jb.172.12.6797-6802.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szittner R., Meighen E. Nucleotide sequence, expression, and properties of luciferase coded by lux genes from a terrestrial bacterium. J Biol Chem. 1990 Sep 25;265(27):16581–16587. [PubMed] [Google Scholar]
- Uemori T., Ishino Y., Fujita K., Asada K., Kato I. Cloning of the DNA polymerase gene of Bacillus caldotenax and characterization of the gene product. J Biochem. 1993 Mar;113(3):401–410. doi: 10.1093/oxfordjournals.jbchem.a124058. [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J. Production of single-stranded plasmid DNA. Methods Enzymol. 1987;153:3–11. doi: 10.1016/0076-6879(87)53044-0. [DOI] [PubMed] [Google Scholar]
- Watanabe H., Hastings J. W. Specificities and properties of three reduced pyridine nucleotide-flavin mononucleotide reductases coupling to bacterial luciferase. Mol Cell Biochem. 1982 May 14;44(3):181–187. doi: 10.1007/BF00238506. [DOI] [PubMed] [Google Scholar]
- Zenno S., Inouye S. Bioluminescent immunoassay using a fusion protein of protein A and the photoprotein aequorin. Biochem Biophys Res Commun. 1990 Aug 31;171(1):169–174. doi: 10.1016/0006-291x(90)91372-y. [DOI] [PubMed] [Google Scholar]
- Zenno S., Saigo K., Kanoh H., Inouye S. Identification of the gene encoding the major NAD(P)H-flavin oxidoreductase of the bioluminescent bacterium Vibrio fischeri ATCC 7744. J Bacteriol. 1994 Jun;176(12):3536–3543. doi: 10.1128/jb.176.12.3536-3543.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]





