Abstract
ATP sulfurylase is a key enzyme in the energy-generating sulfur oxidation pathways of many chemoautotrophic bacteria. The utilization of reduced sulfur compounds to fuel CO2 fixation by the still-uncultured bacterial endosymbionts provides the basis of nutrition in invertebrates, such as the tubeworm Riftia pachyptila, found at deep-sea hydrothermal vents. The symbiont-containing trophosome tissue contains high levels of ATP sulfurylase activity, facilitating the recent purification of the enzyme. The gene encoding the ATP sulfurylase from the Riftia symbiont (sopT) has now been cloned and sequenced by using the partial amino acid sequence of the purified protein. Characterization of the sopT gene has unequivocally shown its bacterial origin. This is the first ATP sulfurylase gene to be cloned and sequenced from a sulfur-oxidizing bacterium. The deduced amino acid sequence was compared to those of ATP sulfurylases reported from organisms which assimilate sulfate, resulting in the discovery that there is substantial homology with the Saccharomyces cerevisiae MET3 gene product but none with the products of the cysDN genes from Escherichia coli nor with the nodP and nodQ genes from Rhizobium meliloti. This and emerging evidence from other sources suggests that E. coli may be atypical, even among prokaryotic sulfate assimilators, in the enzyme it employs for adenosine 5'-phosphosulfate formation. The sopT gene probe also was shown to specifically identify chemoautotrophic bacteria which utilize ATP sulfurylase to oxidize sulfur compounds.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Akagi J. M., Campbell L. L. STUDIES ON THERMOPHILIC SULFATE-REDUCING BACTERIA III. : Adenosine Triphosphate-sulfurylase of Clostridium nigrificans and Desulfovibrio desulfuricans. J Bacteriol. 1962 Dec;84(6):1194–1201. doi: 10.1128/jb.84.6.1194-1201.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brune D. C. Sulfur oxidation by phototrophic bacteria. Biochim Biophys Acta. 1989 Jul 13;975(2):189–221. doi: 10.1016/s0005-2728(89)80251-8. [DOI] [PubMed] [Google Scholar]
- Cavanaugh C. M., Gardiner S. L., Jones M. L., Jannasch H. W., Waterbury J. B. Prokaryotic Cells in the Hydrothermal Vent Tube Worm Riftia pachyptila Jones: Possible Chemoautotrophic Symbionts. Science. 1981 Jul 17;213(4505):340–342. doi: 10.1126/science.213.4505.340. [DOI] [PubMed] [Google Scholar]
- Cherest H., Kerjan P., Surdin-Kerjan Y. The Saccharomyces cerevisiae MET3 gene: nucleotide sequence and relationship of the 5' non-coding region to that of MET25. Mol Gen Genet. 1987 Dec;210(2):307–313. doi: 10.1007/BF00325699. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards D. B., Nelson D. C. DNA-DNA Solution Hybridization Studies of the Bacterial Symbionts of Hydrothermal Vent Tube Worms (Riftia pachyptila and Tevnia jerichonana). Appl Environ Microbiol. 1991 Apr;57(4):1082–1088. doi: 10.1128/aem.57.4.1082-1088.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felbeck H. Chemoautotrophic Potential of the Hydrothermal Vent Tube Worm, Riftia pachyptila Jones (Vestimentifera). Science. 1981 Jul 17;213(4505):336–338. doi: 10.1126/science.213.4505.336. [DOI] [PubMed] [Google Scholar]
- Hawley D. K., McClure W. R. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 1983 Apr 25;11(8):2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyh T. S., Suo Y. GTPase-mediated activation of ATP sulfurylase. J Biol Chem. 1992 Jan 5;267(1):542–545. [PubMed] [Google Scholar]
- Leyh T. S., Taylor J. C., Markham G. D. The sulfate activation locus of Escherichia coli K12: cloning, genetic, and enzymatic characterization. J Biol Chem. 1988 Feb 15;263(5):2409–2416. [PubMed] [Google Scholar]
- Martin R. L., Daley L. A., Lovric Z., Wailes L. M., Renosto F., Segel I. H. The "regulatory" sulfhydryl group of Penicillium chrysogenum ATP sulfurylase. Cooperative ligand binding after SH modification; chemical and thermodynamic properties. J Biol Chem. 1989 Jul 15;264(20):11768–11775. [PubMed] [Google Scholar]
- Nelson D. C., Castenholz R. W. Organic nutrition of Beggiatoa sp. J Bacteriol. 1981 Jul;147(1):236–247. doi: 10.1128/jb.147.1.236-247.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PECK H. D., Jr Enzymatic basis for assimilatory and dissimilatory sulfate reduction. J Bacteriol. 1961 Dec;82:933–939. doi: 10.1128/jb.82.6.933-939.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
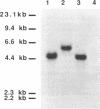
- Renosto F., Martin R. L., Borrell J. L., Nelson D. C., Segel I. H. ATP sulfurylase from trophosome tissue of Riftia pachyptila (hydrothermal vent tube worm). Arch Biochem Biophys. 1991 Oct;290(1):66–78. doi: 10.1016/0003-9861(91)90592-7. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraste M., Sibbald P. R., Wittinghofer A. The P-loop--a common motif in ATP- and GTP-binding proteins. Trends Biochem Sci. 1990 Nov;15(11):430–434. doi: 10.1016/0968-0004(90)90281-f. [DOI] [PubMed] [Google Scholar]
- Schwedock J. S., Long S. R. Rhizobium meliloti genes involved in sulfate activation: the two copies of nodPQ and a new locus, saa. Genetics. 1992 Dec;132(4):899–909. doi: 10.1093/genetics/132.4.899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwedock J., Long S. R. ATP sulphurylase activity of the nodP and nodQ gene products of Rhizobium meliloti. Nature. 1990 Dec 13;348(6302):644–647. doi: 10.1038/348644a0. [DOI] [PubMed] [Google Scholar]
- Segel I. H., Renosto F., Seubert P. A. Sulfate-activating enzymes. Methods Enzymol. 1987;143:334–349. doi: 10.1016/0076-6879(87)43061-9. [DOI] [PubMed] [Google Scholar]