Abstract
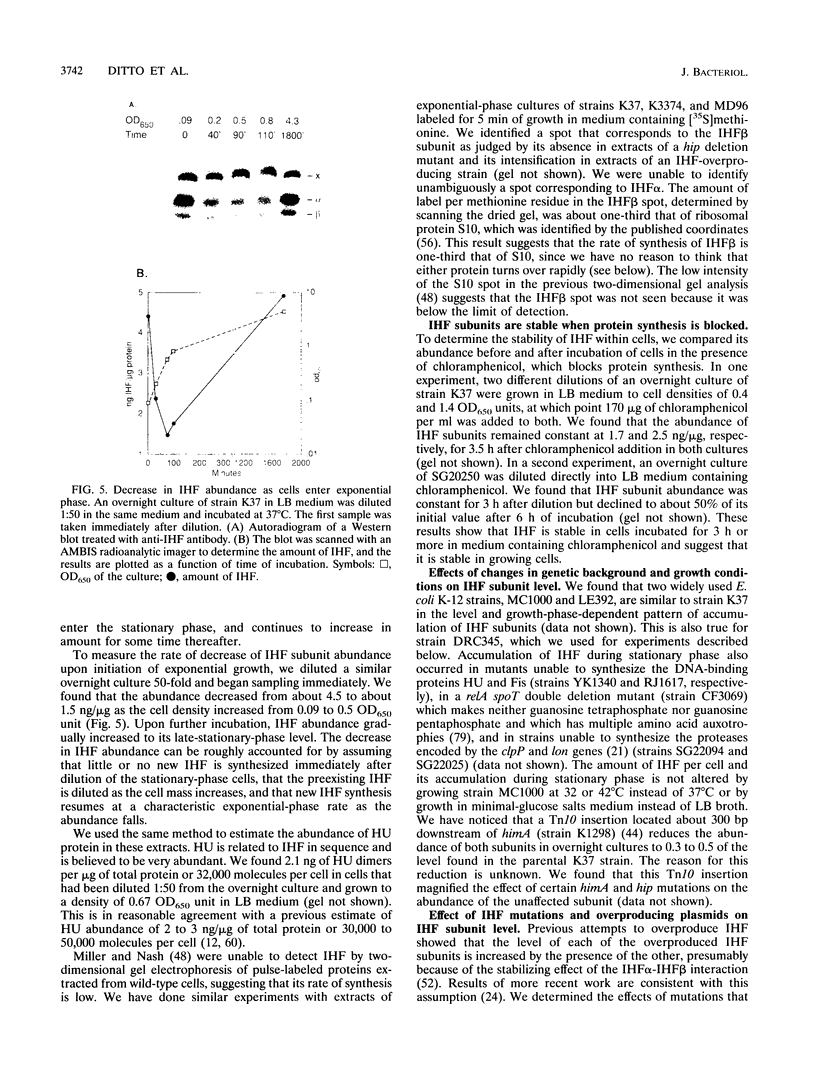
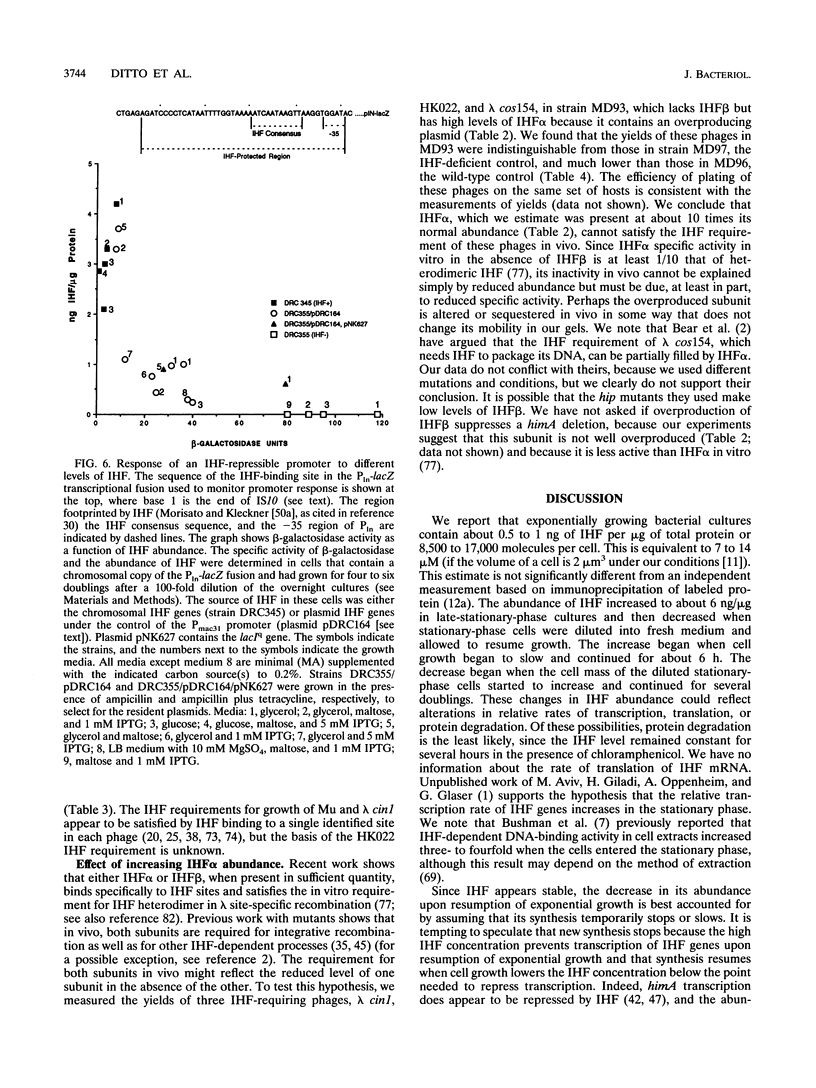
We have measured the intracellular abundance of integration host factor (IHF), a site-specific, heterodimeric DNA-binding protein, in exponential- and stationary-phase cultures of Escherichia coli K-12. Western immunoblot analysis showed that cultures that had been growing exponentially for several generations contained 0.5 to 1.0 ng of IHF subunits per microgram of total protein and that this increased to 5 to 6 ng/microgram in late-stationary-phase cultures. IHF is about one-third to one-half as abundant in exponentially growing cells as HU, a structurally related protein that binds DNA with little or no site specificity. Wild-type IHF is metabolically stable, but deletion mutations that eliminated one subunit reduced the abundance of the other when cells enter stationary phase. We attribute this reduction to the loss of stabilizing interactions between subunits. A mutation that inactivates IHF function but not subunit interaction increased IHF abundance, consistent with results of previous work showing that IHF synthesis is negatively autoregulated. We estimate that steady-state exponential-phase cultures contain about 8,500 to 17,000 IHF dimers per cell, a surprisingly large number for a site-specific DNA-binding protein with a limited number of specific sites. Nevertheless, small reductions in IHF abundance had significant effects on several IHF-dependent functions, suggesting that the wild-type exponential phase level is not in large excess of the minimum required for occupancy of physiologically important IHF-binding sites.
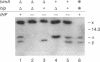
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ball C. A., Osuna R., Ferguson K. C., Johnson R. C. Dramatic changes in Fis levels upon nutrient upshift in Escherichia coli. J Bacteriol. 1992 Dec;174(24):8043–8056. doi: 10.1128/jb.174.24.8043-8056.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bear S. E., Court D. L., Friedman D. I. An accessory role for Escherichia coli integration host factor: characterization of a lambda mutant dependent upon integration host factor for DNA packaging. J Virol. 1984 Dec;52(3):966–972. doi: 10.1128/jvi.52.3.966-972.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benoit R., Böhlen P., Ling N., Briskin A., Esch F., Brazeau P., Ying S. Y., Guillemin R. Presence of somatostatin-28-(1-12) in hypothalamus and pancreas. Proc Natl Acad Sci U S A. 1982 Feb;79(3):917–921. doi: 10.1073/pnas.79.3.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg O. G. The evolutionary selection of DNA base pairs in gene-regulatory binding sites. Proc Natl Acad Sci U S A. 1992 Aug 15;89(16):7501–7505. doi: 10.1073/pnas.89.16.7501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boffini A., Prentki P. Identification of protein binding sites in genomic DNA by two-dimensional gel electrophoresis. Nucleic Acids Res. 1991 Apr 11;19(7):1369–1374. doi: 10.1093/nar/19.7.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnefoy E., Rouvière-Yaniv J. HU and IHF, two homologous histone-like proteins of Escherichia coli, form different protein-DNA complexes with short DNA fragments. EMBO J. 1991 Mar;10(3):687–696. doi: 10.1002/j.1460-2075.1991.tb07998.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushman W., Thompson J. F., Vargas L., Landy A. Control of directionality in lambda site specific recombination. Science. 1985 Nov 22;230(4728):906–911. doi: 10.1126/science.2932798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadaban M. J., Cohen S. N. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J Mol Biol. 1980 Apr;138(2):179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- Dersch P., Schmidt K., Bremer E. Synthesis of the Escherichia coli K-12 nucleoid-associated DNA-binding protein H-NS is subjected to growth-phase control and autoregulation. Mol Microbiol. 1993 May;8(5):875–889. doi: 10.1111/j.1365-2958.1993.tb01634.x. [DOI] [PubMed] [Google Scholar]
- Dijk J., White S. W., Wilson K. S., Appelt K. On the DNA binding protein II from Bacillus stearothermophilus. I. Purification, studies in solution, and crystallization. J Biol Chem. 1983 Mar 25;258(6):4003–4006. [PubMed] [Google Scholar]
- Drlica K., Rouviere-Yaniv J. Histonelike proteins of bacteria. Microbiol Rev. 1987 Sep;51(3):301–319. doi: 10.1128/mr.51.3.301-319.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flamm E. L., Weisberg R. A. Primary structure of the hip gene of Escherichia coli and of its product, the beta subunit of integration host factor. J Mol Biol. 1985 May 25;183(2):117–128. doi: 10.1016/0022-2836(85)90206-2. [DOI] [PubMed] [Google Scholar]
- Freundlich M., Ramani N., Mathew E., Sirko A., Tsui P. The role of integration host factor in gene expression in Escherichia coli. Mol Microbiol. 1992 Sep;6(18):2557–2563. doi: 10.1111/j.1365-2958.1992.tb01432.x. [DOI] [PubMed] [Google Scholar]
- Friedman D. I. Integration host factor: a protein for all reasons. Cell. 1988 Nov 18;55(4):545–554. doi: 10.1016/0092-8674(88)90213-9. [DOI] [PubMed] [Google Scholar]
- Gardner J. F., Nash H. A. Role of Escherichia coli IHF protein in lambda site-specific recombination. A mutational analysis of binding sites. J Mol Biol. 1986 Sep 20;191(2):181–189. doi: 10.1016/0022-2836(86)90255-x. [DOI] [PubMed] [Google Scholar]
- Gober J. W., Shapiro L. A developmentally regulated Caulobacter flagellar promoter is activated by 3' enhancer and IHF binding elements. Mol Biol Cell. 1992 Aug;3(8):913–926. doi: 10.1091/mbc.3.8.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman S. D., Nicholson S. C., Nash H. A. Deformation of DNA during site-specific recombination of bacteriophage lambda: replacement of IHF protein by HU protein or sequence-directed bends. Proc Natl Acad Sci U S A. 1992 Dec 15;89(24):11910–11914. doi: 10.1073/pnas.89.24.11910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich J. A., Schwartz M. L., McClure W. R. Searching for and predicting the activity of sites for DNA binding proteins: compilation and analysis of the binding sites for Escherichia coli integration host factor (IHF). Nucleic Acids Res. 1990 Sep 11;18(17):4993–5000. doi: 10.1093/nar/18.17.4993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goosen N., van de Putte P. Regulation of Mu transposition. I. Localization of the presumed recognition sites for HimD and Ner functions controlling bacteriophage Mu transcription. Gene. 1984 Oct;30(1-3):41–46. doi: 10.1016/0378-1119(84)90103-3. [DOI] [PubMed] [Google Scholar]
- Gottesman S., Maurizi M. R. Regulation by proteolysis: energy-dependent proteases and their targets. Microbiol Rev. 1992 Dec;56(4):592–621. doi: 10.1128/mr.56.4.592-621.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman S., Trisler P., Torres-Cabassa A. Regulation of capsular polysaccharide synthesis in Escherichia coli K-12: characterization of three regulatory genes. J Bacteriol. 1985 Jun;162(3):1111–1119. doi: 10.1128/jb.162.3.1111-1119.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granston A. E., Alessi D. M., Eades L. J., Friedman D. I. A point mutation in the Nul gene of bacteriophage lambda facilitates phage growth in Escherichia coli with himA and gyrB mutations. Mol Gen Genet. 1988 Apr;212(1):149–156. doi: 10.1007/BF00322458. [DOI] [PubMed] [Google Scholar]
- Granston A. E., Nash H. A. Characterization of a set of integration host factor mutants deficient for DNA binding. J Mol Biol. 1993 Nov 5;234(1):45–59. doi: 10.1006/jmbi.1993.1562. [DOI] [PubMed] [Google Scholar]
- Griffo G., Oppenheim A. B., Gottesman M. E. Repression of the lambda pcin promoter by integrative host factor. J Mol Biol. 1989 Sep 5;209(1):55–64. doi: 10.1016/0022-2836(89)90169-1. [DOI] [PubMed] [Google Scholar]
- Groisman E. A., Castilho B. A., Casadaban M. J. In vivo DNA cloning and adjacent gene fusing with a mini-Mu-lac bacteriophage containing a plasmid replicon. Proc Natl Acad Sci U S A. 1984 Mar;81(5):1480–1483. doi: 10.1073/pnas.81.5.1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haluzi H., Goitein D., Koby S., Mendelson I., Teff D., Mengeritsky G., Giladi H., Oppenheim A. B. Genes coding for integration host factor are conserved in gram-negative bacteria. J Bacteriol. 1991 Oct;173(19):6297–6299. doi: 10.1128/jb.173.19.6297-6299.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinton J. C., Santos D. S., Seirafi A., Hulton C. S., Pavitt G. D., Higgins C. F. Expression and mutational analysis of the nucleoid-associated protein H-NS of Salmonella typhimurium. Mol Microbiol. 1992 Aug;6(16):2327–2337. doi: 10.1111/j.1365-2958.1992.tb01408.x. [DOI] [PubMed] [Google Scholar]
- Howe J. G., Hershey J. W. A sensitive immunoblotting method for measuring protein synthesis initiation factor levels in lysates of Escherichia coli. J Biol Chem. 1981 Dec 25;256(24):12836–12839. [PubMed] [Google Scholar]
- Huisman O., Errada P. R., Signon L., Kleckner N. Mutational analysis of IS10's outside end. EMBO J. 1989 Jul;8(7):2101–2109. doi: 10.1002/j.1460-2075.1989.tb03619.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R. C., Ball C. A., Pfeffer D., Simon M. I. Isolation of the gene encoding the Hin recombinational enhancer binding protein. Proc Natl Acad Sci U S A. 1988 May;85(10):3484–3488. doi: 10.1073/pnas.85.10.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P. G., Krah R., Tafuri S. R., Wolffe A. P. DNA gyrase, CS7.4, and the cold shock response in Escherichia coli. J Bacteriol. 1992 Sep;174(18):5798–5802. doi: 10.1128/jb.174.18.5798-5802.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kano Y., Imamoto F. Requirement of integration host factor (IHF) for growth of Escherichia coli deficient in HU protein. Gene. 1990 Apr 30;89(1):133–137. doi: 10.1016/0378-1119(90)90216-e. [DOI] [PubMed] [Google Scholar]
- Kano Y., Ogawa T., Ogura T., Hiraga S., Okazaki T., Imamoto F. Participation of the histone-like protein HU and of IHF in minichromosomal maintenance in Escherichia coli. Gene. 1991 Jul 15;103(1):25–30. doi: 10.1016/0378-1119(91)90386-p. [DOI] [PubMed] [Google Scholar]
- Kikuchi A., Flamm E., Weisberg R. A. An Escherichia coli mutant unable to support site-specific recombination of bacteriophage lambda. J Mol Biol. 1985 May 25;183(2):129–140. doi: 10.1016/0022-2836(85)90207-4. [DOI] [PubMed] [Google Scholar]
- Kolter R., Siegele D. A., Tormo A. The stationary phase of the bacterial life cycle. Annu Rev Microbiol. 1993;47:855–874. doi: 10.1146/annurev.mi.47.100193.004231. [DOI] [PubMed] [Google Scholar]
- Kosturko L. D., Daub E., Murialdo H. The interaction of E. coli integration host factor and lambda cos DNA: multiple complex formation and protein-induced bending. Nucleic Acids Res. 1989 Jan 11;17(1):317–334. doi: 10.1093/nar/17.1.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause H. M., Higgins N. P. Positive and negative regulation of the Mu operator by Mu repressor and Escherichia coli integration host factor. J Biol Chem. 1986 Mar 15;261(8):3744–3752. [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lee E. C., Gumport R. I., Gardner J. F. Genetic analysis of bacteriophage lambda integrase interactions with arm-type attachment site sequences. J Bacteriol. 1990 Mar;172(3):1529–1538. doi: 10.1128/jb.172.3.1529-1538.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z. J., Hillyard D., Higgins P. Nucleotide sequence of the Salmonella typhimurium himA gene. Nucleic Acids Res. 1989 Nov 11;17(21):8880–8880. doi: 10.1093/nar/17.21.8880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mechulam Y., Blanquet S., Fayat G. Dual level control of the Escherichia coli pheST-himA operon expression. tRNA(Phe)-dependent attenuation and transcriptional operator-repressor control by himA and the SOS network. J Mol Biol. 1987 Oct 5;197(3):453–470. doi: 10.1016/0022-2836(87)90558-4. [DOI] [PubMed] [Google Scholar]
- Mendelson I., Gottesman M., Oppenheim A. B. HU and integration host factor function as auxiliary proteins in cleavage of phage lambda cohesive ends by terminase. J Bacteriol. 1991 Mar;173(5):1670–1676. doi: 10.1128/jb.173.5.1670-1676.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller H. I., Friedman D. I. An E. coli gene product required for lambda site-specific recombination. Cell. 1980 Jul;20(3):711–719. doi: 10.1016/0092-8674(80)90317-7. [DOI] [PubMed] [Google Scholar]
- Miller H. I., Kikuchi A., Nash H. A., Weisberg R. A., Friedman D. I. Site-specific recombination of bacteriophage lambda: the role of host gene products. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 2):1121–1126. doi: 10.1101/sqb.1979.043.01.125. [DOI] [PubMed] [Google Scholar]
- Miller H. I., Kirk M., Echols H. SOS induction and autoregulation of the himA gene for site-specific recombination in Escherichia coli. Proc Natl Acad Sci U S A. 1981 Nov;78(11):6754–6758. doi: 10.1073/pnas.78.11.6754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller H. I., Nash H. A. Direct role of the himA gene product in phage lambda integration. Nature. 1981 Apr 9;290(5806):523–526. doi: 10.1038/290523a0. [DOI] [PubMed] [Google Scholar]
- Miller H. I. Primary structure of the himA gene of Escherichia coli: homology with DNA-binding protein HU and association with the phenylalanyl-tRNA synthetase operon. Cold Spring Harb Symp Quant Biol. 1984;49:691–698. doi: 10.1101/sqb.1984.049.01.078. [DOI] [PubMed] [Google Scholar]
- Morisato D., Kleckner N. Tn10 transposition and circle formation in vitro. Cell. 1987 Oct 9;51(1):101–111. doi: 10.1016/0092-8674(87)90014-6. [DOI] [PubMed] [Google Scholar]
- Nash H. A., Robertson C. A., Flamm E., Weisberg R. A., Miller H. I. Overproduction of Escherichia coli integration host factor, a protein with nonidentical subunits. J Bacteriol. 1987 Sep;169(9):4124–4127. doi: 10.1128/jb.169.9.4124-4127.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash H. A., Robertson C. A. Purification and properties of the Escherichia coli protein factor required for lambda integrative recombination. J Biol Chem. 1981 Sep 10;256(17):9246–9253. [PubMed] [Google Scholar]
- Ninnemann O., Koch C., Kahmann R. The E.coli fis promoter is subject to stringent control and autoregulation. EMBO J. 1992 Mar;11(3):1075–1083. doi: 10.1002/j.1460-2075.1992.tb05146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppenheim A. B., Rudd K. E., Mendelson I., Teff D. Integration host factor binds to a unique class of complex repetitive extragenic DNA sequences in Escherichia coli. Mol Microbiol. 1993 Oct;10(1):113–122. doi: 10.1111/j.1365-2958.1993.tb00908.x. [DOI] [PubMed] [Google Scholar]
- Owen-Hughes T. A., Pavitt G. D., Santos D. S., Sidebotham J. M., Hulton C. S., Hinton J. C., Higgins C. F. The chromatin-associated protein H-NS interacts with curved DNA to influence DNA topology and gene expression. Cell. 1992 Oct 16;71(2):255–265. doi: 10.1016/0092-8674(92)90354-f. [DOI] [PubMed] [Google Scholar]
- Prentki P., Chandler M., Galas D. J. Escherichia coli integration host factor bends the DNA at the ends of IS1 and in an insertion hotspot with multiple IHF binding sites. EMBO J. 1987 Aug;6(8):2479–2487. doi: 10.1002/j.1460-2075.1987.tb02529.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimsky S., Spassky A. Sequence determinants for H1 binding on Escherichia coli lac and gal promoters. Biochemistry. 1990 Apr 17;29(15):3765–3771. doi: 10.1021/bi00467a024. [DOI] [PubMed] [Google Scholar]
- Robertson C. A., Nash H. A. Bending of the bacteriophage lambda attachment site by Escherichia coli integration host factor. J Biol Chem. 1988 Mar 15;263(8):3554–3557. [PubMed] [Google Scholar]
- Rouvière-Yaniv J., Kjeldgaard N. O. Native Escherichia coli HU protein is a heterotypic dimer. FEBS Lett. 1979 Oct 15;106(2):297–300. doi: 10.1016/0014-5793(79)80518-9. [DOI] [PubMed] [Google Scholar]
- Schmid M. B., Johnson R. C. Southern revival--news of bacterial chromatin. Prokaryotic chromosomes: structure and function in genome design. Fifth Annual University of Alabama at Birmingham Biochemistry symposium, Panama City, FL, USA, May 8-12, 1991. New Biol. 1991 Oct;3(10):945–950. [PubMed] [Google Scholar]
- Siegele D. A., Kolter R. Life after log. J Bacteriol. 1992 Jan;174(2):345–348. doi: 10.1128/jb.174.2.345-348.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons R. W., Houman F., Kleckner N. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene. 1987;53(1):85–96. doi: 10.1016/0378-1119(87)90095-3. [DOI] [PubMed] [Google Scholar]
- Smith P. K., Krohn R. I., Hermanson G. T., Mallia A. K., Gartner F. H., Provenzano M. D., Fujimoto E. K., Goeke N. M., Olson B. J., Klenk D. C. Measurement of protein using bicinchoninic acid. Anal Biochem. 1985 Oct;150(1):76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- Thompson J. F., Landy A. Empirical estimation of protein-induced DNA bending angles: applications to lambda site-specific recombination complexes. Nucleic Acids Res. 1988 Oct 25;16(20):9687–9705. doi: 10.1093/nar/16.20.9687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J. F., Moitoso de Vargas L., Koch C., Kahmann R., Landy A. Cellular factors couple recombination with growth phase: characterization of a new component in the lambda site-specific recombination pathway. Cell. 1987 Sep 11;50(6):901–908. doi: 10.1016/0092-8674(87)90516-2. [DOI] [PubMed] [Google Scholar]
- Thompson J. F., Waechter-Brulla D., Gumport R. I., Gardner J. F., Moitoso de Vargas L., Landy A. Mutations in an integration host factor-binding site: effect on lambda site-specific recombination and regulatory implications. J Bacteriol. 1986 Dec;168(3):1343–1351. doi: 10.1128/jb.168.3.1343-1351.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobe T., Yoshikawa M., Mizuno T., Sasakawa C. Transcriptional control of the invasion regulatory gene virB of Shigella flexneri: activation by virF and repression by H-NS. J Bacteriol. 1993 Oct;175(19):6142–6149. doi: 10.1128/jb.175.19.6142-6149.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal-Ingigliardi D., Raibaud O. The mac promoters: functional hybrid promoters activated by the malT product and repressed by the lacI product. Nucleic Acids Res. 1985 Feb 25;13(4):1163–1172. doi: 10.1093/nar/13.4.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada M., Kano Y., Ogawa T., Okazaki T., Imamoto F. Construction and characterization of the deletion mutant of hupA and hupB genes in Escherichia coli. J Mol Biol. 1988 Dec 5;204(3):581–591. doi: 10.1016/0022-2836(88)90357-9. [DOI] [PubMed] [Google Scholar]
- Xiao H., Kalman M., Ikehara K., Zemel S., Glaser G., Cashel M. Residual guanosine 3',5'-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. J Biol Chem. 1991 Mar 25;266(9):5980–5990. [PubMed] [Google Scholar]
- Yamada H., Muramatsu S., Mizuno T. An Escherichia coli protein that preferentially binds to sharply curved DNA. J Biochem. 1990 Sep;108(3):420–425. doi: 10.1093/oxfordjournals.jbchem.a123216. [DOI] [PubMed] [Google Scholar]
- Yasuzawa K., Hayashi N., Goshima N., Kohno K., Imamoto F., Kano Y. Histone-like proteins are required for cell growth and constraint of supercoils in DNA. Gene. 1992 Dec 1;122(1):9–15. doi: 10.1016/0378-1119(92)90026-l. [DOI] [PubMed] [Google Scholar]
- Zulianello L., de la Gorgue de Rosny E., van Ulsen P., van de Putte P., Goosen N. The HimA and HimD subunits of integration host factor can specifically bind to DNA as homodimers. EMBO J. 1994 Apr 1;13(7):1534–1540. doi: 10.1002/j.1460-2075.1994.tb06415.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rijn P. A., van de Putte P., Goosen N. Analysis of the IHF binding site in the regulatory region of bacteriophage Mu. Nucleic Acids Res. 1991 Jun 11;19(11):2825–2834. doi: 10.1093/nar/19.11.2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Putte P., Giphart-Gassler M., Goosen N., Goosen T., van Leerdam E. Regulation of integration and replication functions of bacteriophage Mu. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 1):347–353. doi: 10.1101/sqb.1981.045.01.048. [DOI] [PubMed] [Google Scholar]