Abstract
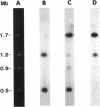
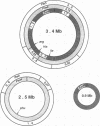
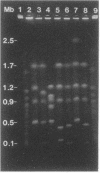
Macrorestriction fragment analysis of DNA from Pseudomonas cepacia 17616, in conjunction with Southern hybridization experiments using junction fragments containing rare restriction enzyme sites as probes, indicated that this bacterium contains three large circular replicons of 3.4, 2.5, and 0.9 megabases (Mb). Inclusion of the 170-kb cryptic plasmid present in this strain gave an overall estimate of genome size of 7 Mb. Other Southern hybridization experiments indicated that the three large replicons contained rRNA genes as well as insertion sequence elements identified previously in this strain. The distribution of SwaI, PacI, and PmeI sites on the three replicons was determined. A derivative of Tn5-751 carrying a SwaI site was used to inactivate and map genes on the 2.5- and 3.4-Mb replicons. Mutants were isolated in which the 2.5- and 0.9-Mb replicons had been reduced in size to 1.8 and 0.65 Mb, respectively. The loss of DNA from the 2.5-Mb replicon was associated with lysine auxotrophy, beta-lactamase deficiency, and failure to utilize ribitol and trehalose as carbon and energy sources. DNA fragments corresponding in size to randomly linearized forms of the different replicons were detected in unrestricted DNA by pulsed-field gel electrophoresis. The results provide a framework for further genetic analysis of strain 17616 and for evaluation of the genomic complexities of other P. cepacia isolates.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allardet-Servent A., Michaux-Charachon S., Jumas-Bilak E., Karayan L., Ramuz M. Presence of one linear and one circular chromosome in the Agrobacterium tumefaciens C58 genome. J Bacteriol. 1993 Dec;175(24):7869–7874. doi: 10.1128/jb.175.24.7869-7874.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballard R. W., Palleroni N. J., Doudoroff M., Stanier R. Y., Mandel M. Taxonomy of the aerobic pseudomonads: Pseudomonas cepacia, P. marginata, P. alliicola and P. caryophylli. J Gen Microbiol. 1970 Feb;60(2):199–214. doi: 10.1099/00221287-60-2-199. [DOI] [PubMed] [Google Scholar]
- Barsomian G., Lessie T. G. IS2 activates the ilvA gene of Pseudomonas cepacia in Escherichia coli. J Bacteriol. 1987 Apr;169(4):1777–1779. doi: 10.1128/jb.169.4.1777-1779.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barsomian G., Lessie T. G. Replicon fusions promoted by insertion sequences on Pseudomonas cepacia plasmid pTGL6. Mol Gen Genet. 1986 Aug;204(2):273–280. doi: 10.1007/BF00425509. [DOI] [PubMed] [Google Scholar]
- Batie C. J., LaHaie E., Ballou D. P. Purification and characterization of phthalate oxygenase and phthalate oxygenase reductase from Pseudomonas cepacia. J Biol Chem. 1987 Feb 5;262(4):1510–1518. [PubMed] [Google Scholar]
- Birnboim H. C. A rapid alkaline extraction method for the isolation of plasmid DNA. Methods Enzymol. 1983;100:243–255. doi: 10.1016/0076-6879(83)00059-2. [DOI] [PubMed] [Google Scholar]
- Byrne A. M., Lessie T. G. Characteristics of IS401, a new member of the IS3 family implicated in plasmid rearrangements in Pseudomonas cepacia. Plasmid. 1994 Mar;31(2):138–147. doi: 10.1006/plas.1994.1015. [DOI] [PubMed] [Google Scholar]
- Cantor C. R., Smith C. L., Mathew M. K. Pulsed-field gel electrophoresis of very large DNA molecules. Annu Rev Biophys Biophys Chem. 1988;17:287–304. doi: 10.1146/annurev.bb.17.060188.001443. [DOI] [PubMed] [Google Scholar]
- Chu G., Vollrath D., Davis R. W. Separation of large DNA molecules by contour-clamped homogeneous electric fields. Science. 1986 Dec 19;234(4783):1582–1585. doi: 10.1126/science.3538420. [DOI] [PubMed] [Google Scholar]
- Ferrante A. A., Lessie T. G. Nucleotide sequence of IS402 from Pseudomonas cepacia. Gene. 1991 Jun 15;102(1):143–144. doi: 10.1016/0378-1119(91)90555-p. [DOI] [PubMed] [Google Scholar]
- Fetzner S., Müller R., Lingens F. Purification and some properties of 2-halobenzoate 1,2-dioxygenase, a two-component enzyme system from Pseudomonas cepacia 2CBS. J Bacteriol. 1992 Jan;174(1):279–290. doi: 10.1128/jb.174.1.279-290.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaffney T. D., Lessie T. G. Insertion-sequence-dependent rearrangements of Pseudomonas cepacia plasmid pTGL1. J Bacteriol. 1987 Jan;169(1):224–230. doi: 10.1128/jb.169.1.224-230.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaubier P., Vega D., Cooke R. Nucleotide sequence of a 2 kb plasmid from Pseudomonas cepacia implicated in the degradation of phenylcarbamate herbicides. DNA Seq. 1992;2(4):269–271. doi: 10.3109/10425179209020813. [DOI] [PubMed] [Google Scholar]
- Grothues D., Tümmler B. New approaches in genome analysis by pulsed-field gel electrophoresis: application to the analysis of Pseudomonas species. Mol Microbiol. 1991 Nov;5(11):2763–2776. doi: 10.1111/j.1365-2958.1991.tb01985.x. [DOI] [PubMed] [Google Scholar]
- Haugland R. A., Sangodkar U. M., Chakrabarty A. M. Repeated sequences including RS1100 from Pseudomonas cepacia AC1100 function as IS elements. Mol Gen Genet. 1990 Jan;220(2):222–228. doi: 10.1007/BF00260485. [DOI] [PubMed] [Google Scholar]
- Haugland R. A., Sangodkar U. M., Sferra P. R., Chakrabarty A. M. Cloning and characterization of a chromosomal DNA region required for growth on 2,4,5-T by Pseudomonas cepacia AC1100. Gene. 1991 Apr;100:65–73. doi: 10.1016/0378-1119(91)90351-b. [DOI] [PubMed] [Google Scholar]
- Holloway B. W., Escuadra M. D., Morgan A. F., Saffery R., Krishnapillai V. The new approaches to whole genome analysis of bacteria. FEMS Microbiol Lett. 1992 Dec 15;100(1-3):101–105. doi: 10.1111/j.1574-6968.1992.tb14026.x. [DOI] [PubMed] [Google Scholar]
- Holloway B. W. Genetics for all bacteria. Annu Rev Microbiol. 1993;47:659–684. doi: 10.1146/annurev.mi.47.100193.003303. [DOI] [PubMed] [Google Scholar]
- Holloway B. W., Morgan A. F. Genome organization in Pseudomonas. Annu Rev Microbiol. 1986;40:79–105. doi: 10.1146/annurev.mi.40.100186.000455. [DOI] [PubMed] [Google Scholar]
- Honeycutt R. J., McClelland M., Sobral B. W. Physical map of the genome of Rhizobium meliloti 1021. J Bacteriol. 1993 Nov;175(21):6945–6952. doi: 10.1128/jb.175.21.6945-6952.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilbane J. J., Chatterjee D. K., Karns J. S., Kellogg S. T., Chakrabarty A. M. Biodegradation of 2,4,5-trichlorophenoxyacetic acid by a pure culture of Pseudomonas cepacia. Appl Environ Microbiol. 1982 Jul;44(1):72–78. doi: 10.1128/aem.44.1.72-78.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krawiec S., Riley M. Organization of the bacterial chromosome. Microbiol Rev. 1990 Dec;54(4):502–539. doi: 10.1128/mr.54.4.502-539.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessie T. G., Phibbs P. V., Jr Alternative pathways of carbohydrate utilization in pseudomonads. Annu Rev Microbiol. 1984;38:359–388. doi: 10.1146/annurev.mi.38.100184.002043. [DOI] [PubMed] [Google Scholar]
- Lessie T. G. The atypical ribosomal RNA complement of Rhodopseudomonas spheroides. J Gen Microbiol. 1965 Jun;39(3):311–320. doi: 10.1099/00221287-39-3-311. [DOI] [PubMed] [Google Scholar]
- Liu S. L., Hessel A., Sanderson K. E. Genomic mapping with I-Ceu I, an intron-encoded endonuclease specific for genes for ribosomal RNA, in Salmonella spp., Escherichia coli, and other bacteria. Proc Natl Acad Sci U S A. 1993 Jul 15;90(14):6874–6878. doi: 10.1073/pnas.90.14.6874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClelland M., Hanish J., Nelson M., Patel Y. KGB: a single buffer for all restriction endonucleases. Nucleic Acids Res. 1988 Jan 11;16(1):364–364. doi: 10.1093/nar/16.1.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClelland M., Jones R., Patel Y., Nelson M. Restriction endonucleases for pulsed field mapping of bacterial genomes. Nucleic Acids Res. 1987 Aug 11;15(15):5985–6005. doi: 10.1093/nar/15.15.5985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaux S., Paillisson J., Carles-Nurit M. J., Bourg G., Allardet-Servent A., Ramuz M. Presence of two independent chromosomes in the Brucella melitensis 16M genome. J Bacteriol. 1993 Feb;175(3):701–705. doi: 10.1128/jb.175.3.701-705.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazawa T., Kimoto M., Abe M. Cloning, sequencing, and transcriptional analysis of the recA gene of Pseudomonas cepacia. Gene. 1990 Sep 28;94(1):83–88. doi: 10.1016/0378-1119(90)90471-3. [DOI] [PubMed] [Google Scholar]
- Proenca R., Niu W. W., Cacalano G., Prince A. The Pseudomonas cepacia 249 chromosomal penicillinase is a member of the AmpC family of chromosomal beta-lactamases. Antimicrob Agents Chemother. 1993 Apr;37(4):667–674. doi: 10.1128/aac.37.4.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratnaningsih E., Dharmsthiti S., Krishnapillai V., Morgan A., Sinclair M., Holloway B. W. A combined physical and genetic map of Pseudomonas aeruginosa PAO. J Gen Microbiol. 1990 Dec;136(12):2351–2357. doi: 10.1099/00221287-136-12-2351. [DOI] [PubMed] [Google Scholar]
- Rella M., Mercenier A., Haas D. Transposon insertion mutagenesis of Pseudomonas aeruginosa with a Tn5 derivative: application to physical mapping of the arc gene cluster. Gene. 1985;33(3):293–303. doi: 10.1016/0378-1119(85)90237-9. [DOI] [PubMed] [Google Scholar]
- Sangodkar U. M., Chapman P. J., Chakrabarty A. M. Cloning, physical mapping and expression of chromosomal genes specifying degradation of the herbicide 2,4,5-T by Pseudomonas cepacia AC1100. Gene. 1988 Nov 30;71(2):267–277. doi: 10.1016/0378-1119(88)90043-1. [DOI] [PubMed] [Google Scholar]
- Scordilis G. E., Ree H., Lessie T. G. Identification of transposable elements which activate gene expression in Pseudomonas cepacia. J Bacteriol. 1987 Jan;169(1):8–13. doi: 10.1128/jb.169.1.8-13.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shields M. S., Montgomery S. O., Cuskey S. M., Chapman P. J., Pritchard P. H. Mutants of Pseudomonas cepacia G4 defective in catabolism of aromatic compounds and trichloroethylene. Appl Environ Microbiol. 1991 Jul;57(7):1935–1941. doi: 10.1128/aem.57.7.1935-1941.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shields M. S., Reagin M. J. Selection of a Pseudomonas cepacia strain constitutive for the degradation of trichloroethylene. Appl Environ Microbiol. 1992 Dec;58(12):3977–3983. doi: 10.1128/aem.58.12.3977-3983.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C. L., Cantor C. R. Purification, specific fragmentation, and separation of large DNA molecules. Methods Enzymol. 1987;155:449–467. doi: 10.1016/0076-6879(87)55030-3. [DOI] [PubMed] [Google Scholar]
- Smith C. L., Condemine G. New approaches for physical mapping of small genomes. J Bacteriol. 1990 Mar;172(3):1167–1172. doi: 10.1128/jb.172.3.1167-1172.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C. L., Kolodner R. D. Mapping of Escherichia coli chromosomal Tn5 and F insertions by pulsed field gel electrophoresis. Genetics. 1988 Jun;119(2):227–236. doi: 10.1093/genetics/119.2.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobral B. W., Honeycutt R. J., Atherly A. G., McClelland M. Electrophoretic separation of the three Rhizobium meliloti replicons. J Bacteriol. 1991 Aug;173(16):5173–5180. doi: 10.1128/jb.173.16.5173-5180.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanier R. Y., Palleroni N. J., Doudoroff M. The aerobic pseudomonads: a taxonomic study. J Gen Microbiol. 1966 May;43(2):159–271. doi: 10.1099/00221287-43-2-159. [DOI] [PubMed] [Google Scholar]
- Suwanto A., Kaplan S. Physical and genetic mapping of the Rhodobacter sphaeroides 2.4.1 genome: genome size, fragment identification, and gene localization. J Bacteriol. 1989 Nov;171(11):5840–5849. doi: 10.1128/jb.171.11.5840-5849.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suwanto A., Kaplan S. Physical and genetic mapping of the Rhodobacter sphaeroides 2.4.1 genome: presence of two unique circular chromosomes. J Bacteriol. 1989 Nov;171(11):5850–5859. doi: 10.1128/jb.171.11.5850-5859.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasek P. H., Frantz B., Sangodkar U. M., Haugland R. A., Chakrabarty A. M. Characterization and nucleotide sequence determination of a repeat element isolated from a 2,4,5-T degrading strain of Pseudomonas cepacia. Gene. 1989;76(2):227–238. doi: 10.1016/0378-1119(89)90163-7. [DOI] [PubMed] [Google Scholar]
- Walker E. M., Arnett J. K., Heath J. D., Norris S. J. Treponema pallidum subsp. pallidum has a single, circular chromosome with a size of approximately 900 kilobase pairs. Infect Immun. 1991 Jul;59(7):2476–2479. doi: 10.1128/iai.59.7.2476-2479.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong K. K., McClelland M. Dissection of the Salmonella typhimurium genome by use of a Tn5 derivative carrying rare restriction sites. J Bacteriol. 1992 Jun;174(11):3807–3811. doi: 10.1128/jb.174.11.3807-3811.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood M. S., Byrne A., Lessie T. G. IS406 and IS407, two gene-activating insertion sequences for Pseudomonas cepacia. Gene. 1991 Aug 30;105(1):101–105. doi: 10.1016/0378-1119(91)90519-h. [DOI] [PubMed] [Google Scholar]
- Wood M. S., Lory C., Lessie T. G. Activation of the lac genes of Tn951 by insertion sequences from Pseudomonas cepacia. J Bacteriol. 1990 Apr;172(4):1719–1724. doi: 10.1128/jb.172.4.1719-1724.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zylstra G. J., Olsen R. H., Ballou D. P. Cloning, expression, and regulation of the Pseudomonas cepacia protocatechuate 3,4-dioxygenase genes. J Bacteriol. 1989 Nov;171(11):5907–5914. doi: 10.1128/jb.171.11.5907-5914.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Bliek A. M., Lincke C. R., Borst P. Circular DNA of 3T6R50 double minute chromosomes. Nucleic Acids Res. 1988 Jun 10;16(11):4841–4851. doi: 10.1093/nar/16.11.4841. [DOI] [PMC free article] [PubMed] [Google Scholar]