Abstract
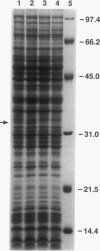
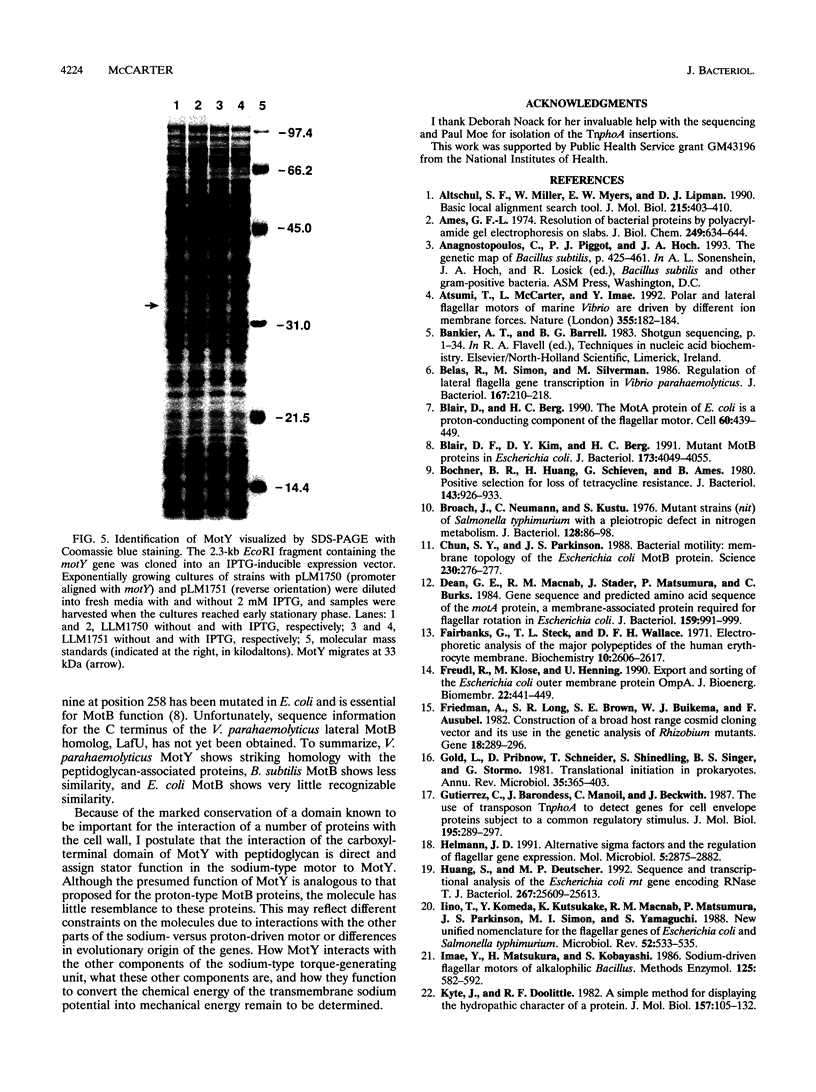
Energy to power the rotation of bacterial flagella can be derived from the proton or sodium transmembrane potential. Until now, genes encoding a bacterial sodium-type flagellar motor have not been defined. A gene, motY, encoding one component of the sodium-type flagellar motor of Vibrio parahaemolyticus was cloned by complementation of a Mot- mutant strain. Sequencing revealed an open reading frame of 879 nucleotides in which a transposon conferring a motility defect mapped. Overexpression of motY in Escherichia coli allowed identification of a product 33 kDa in apparent size on sodium dodecyl sulfate-polyacrylamide gel electrophoresis. This size correlated well with the predicted molecular mass of 33,385 Da. Unlike mot genes identified in other bacteria, localized transposon mutagenesis suggested that the locus was not an extended region containing multiple genes required for swimming motility. Sequencing upstream and downstream of motY confirmed that the gene maps alone and placed it within a locus homologous to the E. coli rnt locus. Although data bank searches failed to reveal significant similarity to known motility components, the carboxyl terminus of MotY showed extensive homology to a number of outer membrane proteins known to interact with peptidoglycan, including OmpA and peptidoglycan-associated lipoproteins. To a limited extent, this domain could also be identified in the Bacillus subtilis MotB protein. This finding suggests that MotY plays the role of a stator in the sodium flagellar motor, stabilizing the force-generating unit through direct interaction with the cell wall.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. Basic local alignment search tool. J Mol Biol. 1990 Oct 5;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Ames G. F. Resolution of bacterial proteins by polyacrylamide gel electrophoresis on slabs. Membrane, soluble, and periplasmic fractions. J Biol Chem. 1974 Jan 25;249(2):634–644. [PubMed] [Google Scholar]
- Atsumi T., McCarter L., Imae Y. Polar and lateral flagellar motors of marine Vibrio are driven by different ion-motive forces. Nature. 1992 Jan 9;355(6356):182–184. doi: 10.1038/355182a0. [DOI] [PubMed] [Google Scholar]
- Belas R., Simon M., Silverman M. Regulation of lateral flagella gene transcription in Vibrio parahaemolyticus. J Bacteriol. 1986 Jul;167(1):210–218. doi: 10.1128/jb.167.1.210-218.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair D. F., Berg H. C. The MotA protein of E. coli is a proton-conducting component of the flagellar motor. Cell. 1990 Feb 9;60(3):439–449. doi: 10.1016/0092-8674(90)90595-6. [DOI] [PubMed] [Google Scholar]
- Blair D. F., Kim D. Y., Berg H. C. Mutant MotB proteins in Escherichia coli. J Bacteriol. 1991 Jul;173(13):4049–4055. doi: 10.1128/jb.173.13.4049-4055.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochner B. R., Huang H. C., Schieven G. L., Ames B. N. Positive selection for loss of tetracycline resistance. J Bacteriol. 1980 Aug;143(2):926–933. doi: 10.1128/jb.143.2.926-933.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broach J., Neumann C., Kustu S. Mutant strains (nit) of Salmonella typhimurium with a pleiotropic defect in nitrogen metabolism. J Bacteriol. 1976 Oct;128(1):86–98. doi: 10.1128/jb.128.1.86-98.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun S. Y., Parkinson J. S. Bacterial motility: membrane topology of the Escherichia coli MotB protein. Science. 1988 Jan 15;239(4837):276–278. doi: 10.1126/science.2447650. [DOI] [PubMed] [Google Scholar]
- Dean G. E., Macnab R. M., Stader J., Matsumura P., Burks C. Gene sequence and predicted amino acid sequence of the motA protein, a membrane-associated protein required for flagellar rotation in Escherichia coli. J Bacteriol. 1984 Sep;159(3):991–999. doi: 10.1128/jb.159.3.991-999.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairbanks G., Steck T. L., Wallach D. F. Electrophoretic analysis of the major polypeptides of the human erythrocyte membrane. Biochemistry. 1971 Jun 22;10(13):2606–2617. doi: 10.1021/bi00789a030. [DOI] [PubMed] [Google Scholar]
- Freudl R., Klose M., Henning U. Export and sorting of the Escherichia coli outer membrane protein OmpA. J Bioenerg Biomembr. 1990 Jun;22(3):441–449. doi: 10.1007/BF00763176. [DOI] [PubMed] [Google Scholar]
- Friedman A. M., Long S. R., Brown S. E., Buikema W. J., Ausubel F. M. Construction of a broad host range cosmid cloning vector and its use in the genetic analysis of Rhizobium mutants. Gene. 1982 Jun;18(3):289–296. doi: 10.1016/0378-1119(82)90167-6. [DOI] [PubMed] [Google Scholar]
- Gold L., Pribnow D., Schneider T., Shinedling S., Singer B. S., Stormo G. Translational initiation in prokaryotes. Annu Rev Microbiol. 1981;35:365–403. doi: 10.1146/annurev.mi.35.100181.002053. [DOI] [PubMed] [Google Scholar]
- Gutierrez C., Barondess J., Manoil C., Beckwith J. The use of transposon TnphoA to detect genes for cell envelope proteins subject to a common regulatory stimulus. Analysis of osmotically regulated genes in Escherichia coli. J Mol Biol. 1987 May 20;195(2):289–297. doi: 10.1016/0022-2836(87)90650-4. [DOI] [PubMed] [Google Scholar]
- Helmann J. D. Alternative sigma factors and the regulation of flagellar gene expression. Mol Microbiol. 1991 Dec;5(12):2875–2882. doi: 10.1111/j.1365-2958.1991.tb01847.x. [DOI] [PubMed] [Google Scholar]
- Huang S., Deutscher M. P. Sequence and transcriptional analysis of the Escherichia coli rnt gene encoding RNase T. J Biol Chem. 1992 Dec 15;267(35):25609–25613. [PubMed] [Google Scholar]
- Iino T., Komeda Y., Kutsukake K., Macnab R. M., Matsumura P., Parkinson J. S., Simon M. I., Yamaguchi S. New unified nomenclature for the flagellar genes of Escherichia coli and Salmonella typhimurium. Microbiol Rev. 1988 Dec;52(4):533–535. doi: 10.1128/mr.52.4.533-535.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imae Y., Matsukura H., Kobayasi S. Sodium-driven flagellar motors of alkalophilic Bacillus. Methods Enzymol. 1986;125:582–592. doi: 10.1016/s0076-6879(86)25047-8. [DOI] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Larsen S. H., Adler J., Gargus J. J., Hogg R. W. Chemomechanical coupling without ATP: the source of energy for motility and chemotaxis in bacteria. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1239–1243. doi: 10.1073/pnas.71.4.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazzaroni J. C., Portalier R. The excC gene of Escherichia coli K-12 required for cell envelope integrity encodes the peptidoglycan-associated lipoprotein (PAL). Mol Microbiol. 1992 Mar;6(6):735–742. doi: 10.1111/j.1365-2958.1992.tb01523.x. [DOI] [PubMed] [Google Scholar]
- Macnab R. M. Proton-driven bacterial flagellar motor. Methods Enzymol. 1986;125:563–581. doi: 10.1016/s0076-6879(86)25046-6. [DOI] [PubMed] [Google Scholar]
- Manoil C., Beckwith J. TnphoA: a transposon probe for protein export signals. Proc Natl Acad Sci U S A. 1985 Dec;82(23):8129–8133. doi: 10.1073/pnas.82.23.8129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M., Showalter R., Silverman M. Identification of a locus controlling expression of luminescence genes in Vibrio harveyi. J Bacteriol. 1989 May;171(5):2406–2414. doi: 10.1128/jb.171.5.2406-2414.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarter L. L., Silverman M. Phosphate regulation of gene expression in Vibrio parahaemolyticus. J Bacteriol. 1987 Aug;169(8):3441–3449. doi: 10.1128/jb.169.8.3441-3449.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarter L. L., Wright M. E. Identification of genes encoding components of the swarmer cell flagellar motor and propeller and a sigma factor controlling differentiation of Vibrio parahaemolyticus. J Bacteriol. 1993 Jun;175(11):3361–3371. doi: 10.1128/jb.175.11.3361-3371.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarter L., Hilmen M., Silverman M. Flagellar dynamometer controls swarmer cell differentiation of V. parahaemolyticus. Cell. 1988 Jul 29;54(3):345–351. doi: 10.1016/0092-8674(88)90197-3. [DOI] [PubMed] [Google Scholar]
- McCarter L., Silverman M. Surface-induced swarmer cell differentiation of Vibrio parahaemolyticus. Mol Microbiol. 1990 Jul;4(7):1057–1062. doi: 10.1111/j.1365-2958.1990.tb00678.x. [DOI] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Mirel D. B., Lustre V. M., Chamberlin M. J. An operon of Bacillus subtilis motility genes transcribed by the sigma D form of RNA polymerase. J Bacteriol. 1992 Jul;174(13):4197–4204. doi: 10.1128/jb.174.13.4197-4204.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman M., Showalter R., McCarter L. Genetic analysis in vibrio. Methods Enzymol. 1991;204:515–536. doi: 10.1016/0076-6879(91)04026-k. [DOI] [PubMed] [Google Scholar]