Abstract
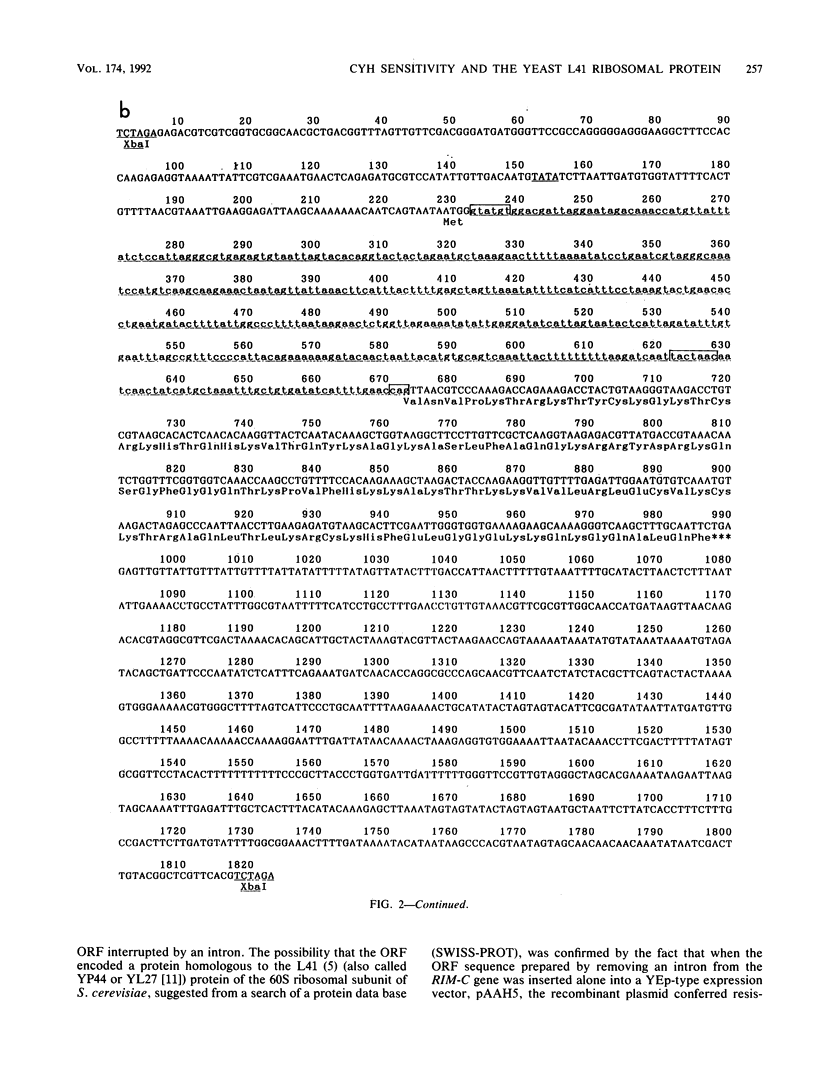
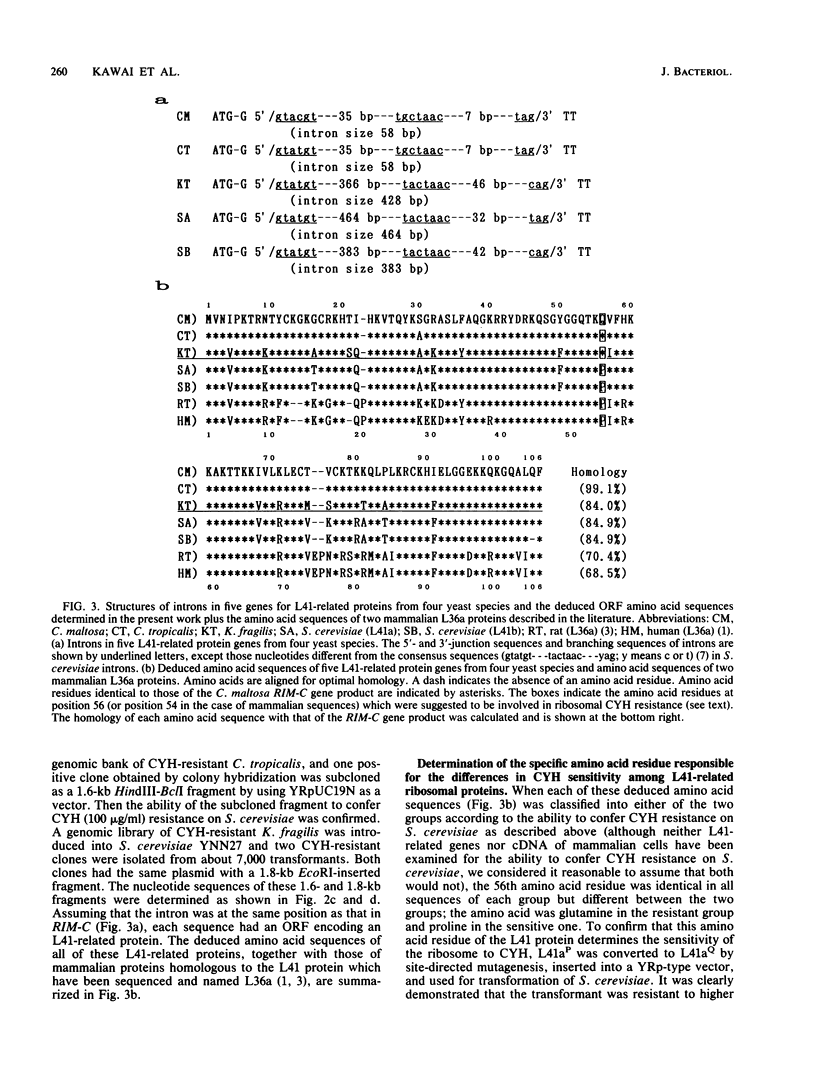
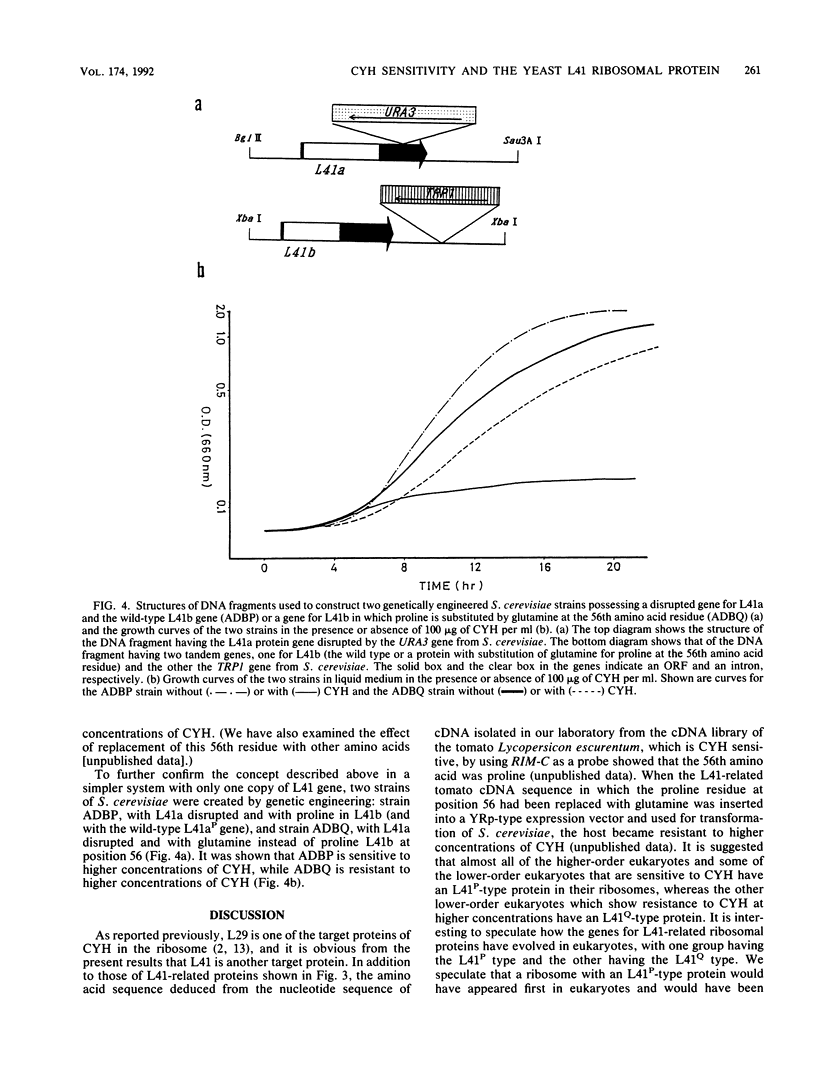
Cycloheximide is one of the antibiotics that inhibit protein synthesis in most eukaryotic cells. We have found that a yeast, Candida maltosa, is resistant to the drug because it possesses a cycloheximide-resistant ribosome, and we have isolated the gene responsible for this. In this study, we sequenced this gene and found that the gene encodes a protein homologous to the L41 ribosomal protein of Saccharomyces cerevisiae, whose amino acid sequence has already been reported. Two genes for L41 protein, named L41a and L41b, independently present in the genome of S. cerevisiae, were isolated. L41-related genes were also isolated from a few other yeast species. Each of these genes has an intron at the same site of the open reading frame. Comparison of their deduced amino acid sequences and their ability to confer cycloheximide resistance to S. cerevisiae, when introduced in a high-copy-number plasmid, suggested that the 56th amino acid residue of the L41 protein determines the sensitivity of the ribosome to cycloheximide; the amino acid is glutamine in the resistant ribosome, whereas that in the sensitive ribosome is proline. This was confirmed by constructing a cycloheximide-resistant strain of S. cerevisiae having a disrupted L41a gene and an L41b gene with a substitution of the glutamine codon for the proline codon.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Davies M. S., Henney A., Ward W. H., Craig R. K. Characterisation of an mRNA encoding a human ribosomal protein homologous to the yeast L44 ribosomal protein. Gene. 1986;45(2):183–191. doi: 10.1016/0378-1119(86)90253-2. [DOI] [PubMed] [Google Scholar]
- Fried H. M., Warner J. R. Molecular cloning and analysis of yeast gene for cycloheximide resistance and ribosomal protein L29. Nucleic Acids Res. 1982 May 25;10(10):3133–3148. doi: 10.1093/nar/10.10.3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher M. J., Chan Y. L., Lin A., Wool I. G. Primary structure of rat ribosomal protein L36a. DNA. 1988 May;7(4):269–273. doi: 10.1089/dna.1988.7.269. [DOI] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh T., Wittmann-Liebold B. The primary structure of protein 44 from the large subunit of yeast ribosomes. FEBS Lett. 1978 Dec 15;96(2):399–402. doi: 10.1016/0014-5793(78)80447-5. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langford C. J., Klinz F. J., Donath C., Gallwitz D. Point mutations identify the conserved, intron-contained TACTAAC box as an essential splicing signal sequence in yeast. Cell. 1984 Mar;36(3):645–653. doi: 10.1016/0092-8674(84)90344-1. [DOI] [PubMed] [Google Scholar]
- Mager W. H. Control of ribosomal protein gene expression. Biochim Biophys Acta. 1988 Jan 25;949(1):1–15. doi: 10.1016/0167-4781(88)90048-6. [DOI] [PubMed] [Google Scholar]
- Mann C., Buhler J. M., Treich I., Sentenac A. RPC40, a unique gene for a subunit shared between yeast RNA polymerases A and C. Cell. 1987 Feb 27;48(4):627–637. doi: 10.1016/0092-8674(87)90241-8. [DOI] [PubMed] [Google Scholar]
- Ohkuma M., Tanimoto T., Yano K., Takagi M. CYP52 (cytochrome P450alk) multigene family in Candida maltosa: molecular cloning and nucleotide sequence of the two tandemly arranged genes. DNA Cell Biol. 1991 May;10(4):271–282. doi: 10.1089/dna.1991.10.271. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K., Stinchcomb D. T., Scherer S., Davis R. W. High-frequency transformation of yeast: autonomous replication of hybrid DNA molecules. Proc Natl Acad Sci U S A. 1979 Mar;76(3):1035–1039. doi: 10.1073/pnas.76.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi M., Kawai S., Shibuya I., Miyazaki M., Yano K. Cloning in Saccharomyces cerevisiae of a cycloheximide resistance gene from the Candida maltosa genome which modifies ribosomes. J Bacteriol. 1986 Oct;168(1):417–419. doi: 10.1128/jb.168.1.417-419.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]



