Abstract
A protein carboxyl methyltransferase (EC 2.1.1.77) that recognizes age-damaged proteins for potential repair or degradation reactions has been found in all vertebrate tissues and cells examined to date. This enzyme catalyzes the transfer of methyl groups from S-adenosylmethionine to the carboxyl groups of D-aspartyl or L-isoaspartyl residues that are formed spontaneously from normal L-aspartyl and L-asparaginyl residues. A similar methyltransferase has been found in two bacterial species, Escherichia coli and Salmonella typhimurium, suggesting that this enzyme performs an essential function in all cells. In this study, we show that this enzyme is present in cytosolic extracts of six additional members of the alpha and gamma subdivisions of the purple bacteria: Pseudomonas aeruginosa (gamma), Rhodobacter sphaeroides (alpha), and the gamma enteric species Klebsiella pneumoniae, Enterobacter aerogenes, Proteus vulgaris, and Serratia marcescens. DNA probes from the E. coli methyltransferase gene hybridized only to the chromosomal DNA of the enteric species. Interestingly, no activity was found in the plant pathogen Erwinia chrysanthemi, a member of the enteric family, nor in Rhizobium meliloti or Rhodopseudomonas palustris, two members of the alpha subdivision. Additionally, we could not detect activity in the four gram-positive species Bacillus subtilis, B. stearothermophilus, Lactobacillus casei, and Streptomyces griseus. The absence of enzyme activity was not due to the presence of inhibitors in the extracts. These results suggest that many cells may not have the enzymatic machinery to recognize abnormal aspartyl residues by methylation reactions. Since the nonenzymatic degradation reactions that generate these residues occur in all cells, other pathways may be present in nature to ensure that these types of altered proteins do not accumulate and interfere with normal cellular physiology.
Full text
PDF

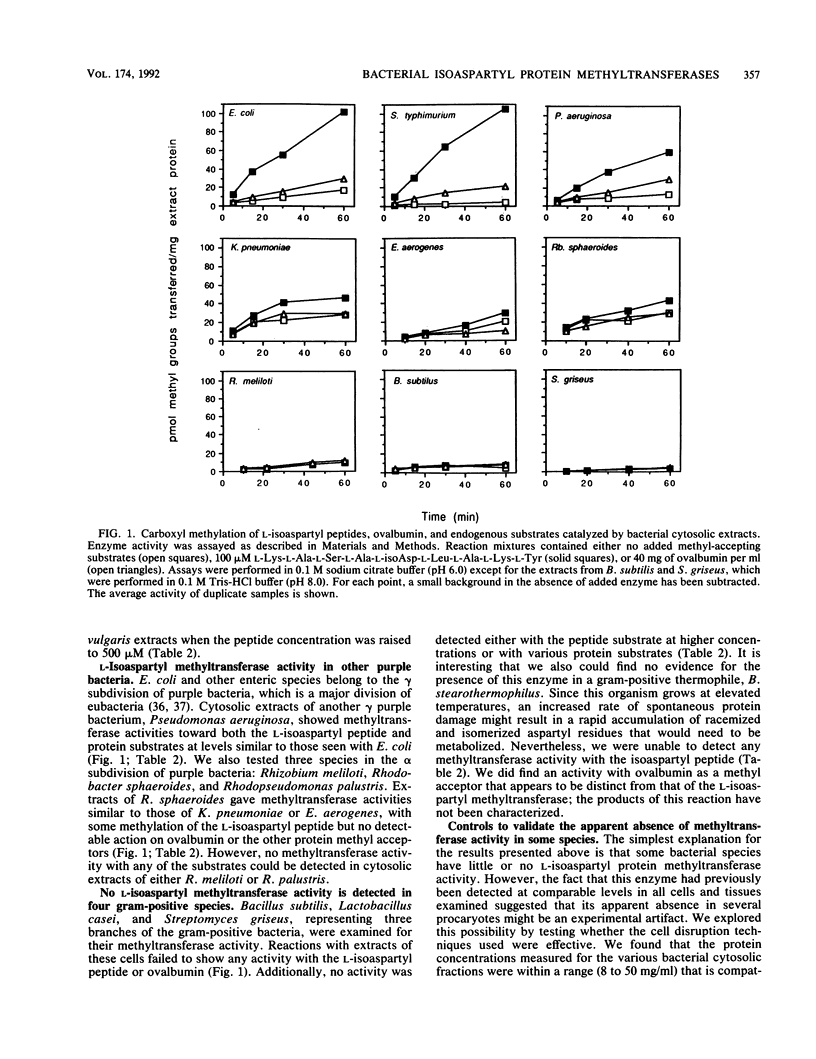




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ahmad S., Johnson J. L., Jensen R. A. The recent evolutionary origin of the phenylalanine-sensitive isozyme of 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase in the enteric lineage of bacteria. J Mol Evol. 1987;25(2):159–167. doi: 10.1007/BF02101758. [DOI] [PubMed] [Google Scholar]
- Barten D. M., O'Dea R. F. The function of protein carboxylmethyltransferase in eucaryotic cells. Life Sci. 1990;47(3):181–194. doi: 10.1016/0024-3205(90)90319-m. [DOI] [PubMed] [Google Scholar]
- Clarke S. Protein carboxyl methyltransferases: two distinct classes of enzymes. Annu Rev Biochem. 1985;54:479–506. doi: 10.1146/annurev.bi.54.070185.002403. [DOI] [PubMed] [Google Scholar]
- Clarke S., Sparrow K., Panasenko S., Koshland D. E., Jr In vitro methylation of bacterial chemotaxis proteins: characterization of protein methyltransferase activity in crude extracts of Salmonella typhimurium. J Supramol Struct. 1980;13(3):315–328. doi: 10.1002/jss.400130305. [DOI] [PubMed] [Google Scholar]
- Doolittle R. F., Feng D. F., Johnson M. S., McClure M. A. Relationships of human protein sequences to those of other organisms. Cold Spring Harb Symp Quant Biol. 1986;51(Pt 1):447–455. doi: 10.1101/sqb.1986.051.01.054. [DOI] [PubMed] [Google Scholar]
- Fox G. E., Stackebrandt E., Hespell R. B., Gibson J., Maniloff J., Dyer T. A., Wolfe R. S., Balch W. E., Tanner R. S., Magrum L. J. The phylogeny of prokaryotes. Science. 1980 Jul 25;209(4455):457–463. doi: 10.1126/science.6771870. [DOI] [PubMed] [Google Scholar]
- Fu J. C., Ding L., Clarke S. Purification, gene cloning, and sequence analysis of an L-isoaspartyl protein carboxyl methyltransferase from Escherichia coli. J Biol Chem. 1991 Aug 5;266(22):14562–14572. [PubMed] [Google Scholar]
- Galletti P., Ciardiello A., Ingrosso D., Di Donato A., D'Alessio G. Repair of isopeptide bonds by protein carboxyl O-methyltransferase: seminal ribonuclease as a model system. Biochemistry. 1988 Mar 8;27(5):1752–1757. doi: 10.1021/bi00405a055. [DOI] [PubMed] [Google Scholar]
- Galletti P., de Rosa M., Gambacorta A., Manna C., Festinese R., Zappia V. Protein methylation in Caldariella acidophila, an extreme thermo-acidophilic archaebacterium. FEBS Lett. 1981 Feb 9;124(1):62–66. doi: 10.1016/0014-5793(81)80054-3. [DOI] [PubMed] [Google Scholar]
- Geiger T., Clarke S. Deamidation, isomerization, and racemization at asparaginyl and aspartyl residues in peptides. Succinimide-linked reactions that contribute to protein degradation. J Biol Chem. 1987 Jan 15;262(2):785–794. [PubMed] [Google Scholar]
- George-Nascimento C., Lowenson J., Borissenko M., Calderón M., Medina-Selby A., Kuo J., Clarke S., Randolph A. Replacement of a labile aspartyl residue increases the stability of human epidermal growth factor. Biochemistry. 1990 Oct 16;29(41):9584–9591. doi: 10.1021/bi00493a012. [DOI] [PubMed] [Google Scholar]
- Gilbert J. M., Fowler A., Bleibaum J., Clarke S. Purification of homologous protein carboxyl methyltransferase isozymes from human and bovine erythrocytes. Biochemistry. 1988 Jul 12;27(14):5227–5233. doi: 10.1021/bi00414a042. [DOI] [PubMed] [Google Scholar]
- Haklai R., Kloog Y. Purification and characterization of protein carboxyl methyltransferase from Torpedo ocellata electric organ. Biochemistry. 1987 Jul 14;26(14):4200–4206. doi: 10.1021/bi00388a004. [DOI] [PubMed] [Google Scholar]
- Ingrosso D., Fowler A. V., Bleibaum J., Clarke S. Sequence of the D-aspartyl/L-isoaspartyl protein methyltransferase from human erythrocytes. Common sequence motifs for protein, DNA, RNA, and small molecule S-adenosylmethionine-dependent methyltransferases. J Biol Chem. 1989 Nov 25;264(33):20131–20139. [PubMed] [Google Scholar]
- Johnson B. A., Langmack E. L., Aswad D. W. Partial repair of deamidation-damaged calmodulin by protein carboxyl methyltransferase. J Biol Chem. 1987 Sep 5;262(25):12283–12287. [PubMed] [Google Scholar]
- Johnson B. A., Murray E. D., Jr, Clarke S., Glass D. B., Aswad D. W. Protein carboxyl methyltransferase facilitates conversion of atypical L-isoaspartyl peptides to normal L-aspartyl peptides. J Biol Chem. 1987 Apr 25;262(12):5622–5629. [PubMed] [Google Scholar]
- Kim S., Lew B., Chang F. N. Enzymatic methyl esterification of Escherichia coli ribosomal proteins. J Bacteriol. 1977 May;130(2):839–845. doi: 10.1128/jb.130.2.839-845.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowenson J. D., Clarke S. Structural elements affecting the recognition of L-isoaspartyl residues by the L-isoaspartyl/D-aspartyl protein methyltransferase. Implications for the repair hypothesis. J Biol Chem. 1991 Oct 15;266(29):19396–19406. [PubMed] [Google Scholar]
- Matin A., Auger E. A., Blum P. H., Schultz J. E. Genetic basis of starvation survival in nondifferentiating bacteria. Annu Rev Microbiol. 1989;43:293–316. doi: 10.1146/annurev.mi.43.100189.001453. [DOI] [PubMed] [Google Scholar]
- McFadden P. N., Clarke S. Conversion of isoaspartyl peptides to normal peptides: implications for the cellular repair of damaged proteins. Proc Natl Acad Sci U S A. 1987 May;84(9):2595–2599. doi: 10.1073/pnas.84.9.2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momand J., Clarke S. Rapid degradation of D- and L-succinimide-containing peptides by a post-proline endopeptidase from human erythrocytes. Biochemistry. 1987 Dec 1;26(24):7798–7805. doi: 10.1021/bi00398a040. [DOI] [PubMed] [Google Scholar]
- Mutoh N., Simon M. I. Nucleotide sequence corresponding to five chemotaxis genes in Escherichia coli. J Bacteriol. 1986 Jan;165(1):161–166. doi: 10.1128/jb.165.1.161-166.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor C. M., Clarke S. Specific recognition of altered polypeptides by widely distributed methyltransferases. Biochem Biophys Res Commun. 1985 Nov 15;132(3):1144–1150. doi: 10.1016/0006-291x(85)91926-6. [DOI] [PubMed] [Google Scholar]
- O'Connor C. M. Regulation and subcellular distribution of a protein methyltransferase and its damaged aspartyl substrate sites in developing Xenopus oocytes. J Biol Chem. 1987 Jul 25;262(21):10398–10403. [PubMed] [Google Scholar]
- Roszak D. B., Colwell R. R. Survival strategies of bacteria in the natural environment. Microbiol Rev. 1987 Sep;51(3):365–379. doi: 10.1128/mr.51.3.365-379.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simms S. A., Stock A. M., Stock J. B. Purification and characterization of the S-adenosylmethionine:glutamyl methyltransferase that modifies membrane chemoreceptor proteins in bacteria. J Biol Chem. 1987 Jun 25;262(18):8537–8543. [PubMed] [Google Scholar]
- Stackebrandt E., Ludwig W., Weizenegger M., Dorn S., McGill T. J., Fox G. E., Woese C. R., Schubert W., Schleifer K. H. Comparative 16S rRNA oligonucleotide analyses and murein types of round-spore-forming bacilli and non-spore-forming relatives. J Gen Microbiol. 1987 Sep;133(9):2523–2529. doi: 10.1099/00221287-133-9-2523. [DOI] [PubMed] [Google Scholar]
- Stephenson R. C., Clarke S. Succinimide formation from aspartyl and asparaginyl peptides as a model for the spontaneous degradation of proteins. J Biol Chem. 1989 Apr 15;264(11):6164–6170. [PubMed] [Google Scholar]
- Syvanen M. Molecular clocks and evolutionary relationships: possible distortions due to horizontal gene flow. J Mol Evol. 1987;26(1-2):16–23. doi: 10.1007/BF02111278. [DOI] [PubMed] [Google Scholar]
- Weisman L. S., Ballou C. E. Methanol production by Mycobacterium smegmatis. J Bacteriol. 1988 Mar;170(3):1393–1395. doi: 10.1128/jb.170.3.1393-1395.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R. Bacterial evolution. Microbiol Rev. 1987 Jun;51(2):221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]


