Abstract
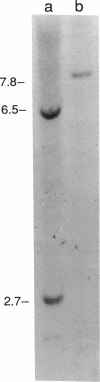
DNA sequence analysis has revealed that an unidentified open reading frame (ufr1) is present immediately preceding the aroB gene of Escherichia coli. The predicted protein product of urf1 contains a consensus ATP-binding-site sequence and shows 34% amino acid homology to shikimate kinase II in a 97-amino-acid region. Inactivation of urf1 by insertion of an antibiotic resistance gene had a polar effect on aroB, indicating that these two genes constitute a transcriptional unit. The auxotrophic requirements of a strain mutant for both urf1 and aroL (encoding shikimate kinase II) are consistent with shikimate kinase deficiency. We propose that urf1 encodes shikimate kinase I and that it be designated aroK.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bachmann B. J. Linkage map of Escherichia coli K-12, edition 8. Microbiol Rev. 1990 Jun;54(2):130–197. doi: 10.1128/mr.54.2.130-197.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlyn M. B., Giles N. H. Organization of enzymes in the polyaromatic synthetic pathway: separability in bacteria. J Bacteriol. 1969 Jul;99(1):222–230. doi: 10.1128/jb.99.1.222-230.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFeyter R. C., Pittard J. Genetic and molecular analysis of aroL, the gene for shikimate kinase II in Escherichia coli K-12. J Bacteriol. 1986 Jan;165(1):226–232. doi: 10.1128/jb.165.1.226-232.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFeyter R. C., Pittard J. Purification and properties of shikimate kinase II from Escherichia coli K-12. J Bacteriol. 1986 Jan;165(1):331–333. doi: 10.1128/jb.165.1.331-333.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ely B., Pittard J. Aromatic amino acid biosynthesis: regulation of shikimate kinase in Escherichia coli K-12. J Bacteriol. 1979 Jun;138(3):933–943. doi: 10.1128/jb.138.3.933-943.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HOWARD-FLANDERS P., SIMSON E., THERIOT L. A LOCUS THAT CONTROLS FILAMENT FORMATION AND SENSITIVITY TO RADIATION IN ESCHERICHIA COLI K-12. Genetics. 1964 Feb;49:237–246. doi: 10.1093/genetics/49.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LURIA S. E., BURROUS J. W. Hybridization between Escherichia coli and Shigella. J Bacteriol. 1957 Oct;74(4):461–476. doi: 10.1128/jb.74.4.461-476.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipman D. J., Pearson W. R. Rapid and sensitive protein similarity searches. Science. 1985 Mar 22;227(4693):1435–1441. doi: 10.1126/science.2983426. [DOI] [PubMed] [Google Scholar]
- Low B. Rapid mapping of conditional and auxotrophic mutations in Escherichia coli K-12. J Bacteriol. 1973 Feb;113(2):798–812. doi: 10.1128/jb.113.2.798-812.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinus M. G. Location of DNA methylation genes on the Escherichia coli K-12 genetic map. Mol Gen Genet. 1973 Dec 14;127(1):47–55. doi: 10.1007/BF00267782. [DOI] [PubMed] [Google Scholar]
- Marinus M. G., Loutit J. S. Regulation of isoleucine-valine biosynthesis in Pseudomonas aeruginosa. I. Characterisation and mapping of mutants. Genetics. 1969 Nov;63(3):547–556. doi: 10.1093/genetics/63.3.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar G., Coggins J. R. The complete amino acid sequence of 3-dehydroquinate synthase of Escherichia coli K12. FEBS Lett. 1986 May 5;200(1):11–17. doi: 10.1016/0014-5793(86)80501-4. [DOI] [PubMed] [Google Scholar]
- Parker B., Marinus M. G. A simple and rapid method to obtain substitution mutations in Escherichia coli: isolation of a dam deletion/insertion mutation. Gene. 1988 Dec 20;73(2):531–535. doi: 10.1016/0378-1119(88)90517-3. [DOI] [PubMed] [Google Scholar]
- Pittard J., Wallace B. J. Distribution and function of genes concerned with aromatic biosynthesis in Escherichia coli. J Bacteriol. 1966 Apr;91(4):1494–1508. doi: 10.1128/jb.91.4.1494-1508.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen L. J., Møller P. L., Atlung T. Carbon metabolism regulates expression of the pfl (pyruvate formate-lyase) gene in Escherichia coli. J Bacteriol. 1991 Oct;173(20):6390–6397. doi: 10.1128/jb.173.20.6390-6397.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rewinski C., Marinus M. G. Mutation spectrum in Escherichia coli DNA mismatch repair deficient (mutH) strain. Nucleic Acids Res. 1987 Oct 26;15(20):8205–8215. doi: 10.1093/nar/15.20.8205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Walker J. E., Saraste M., Runswick M. J., Gay N. J. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1(8):945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Pittard J. Molecular analysis of the regulatory region of the Escherichia coli K-12 tyrB gene. J Bacteriol. 1987 Oct;169(10):4710–4715. doi: 10.1128/jb.169.10.4710-4715.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]