Abstract
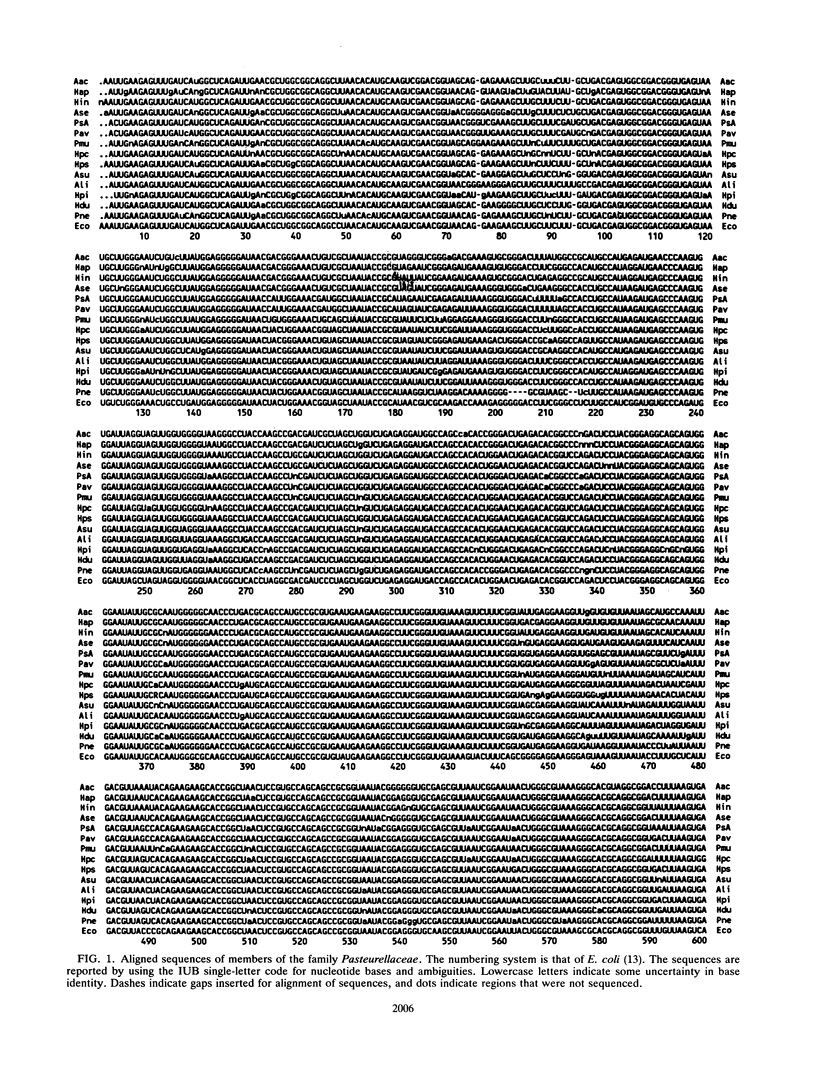
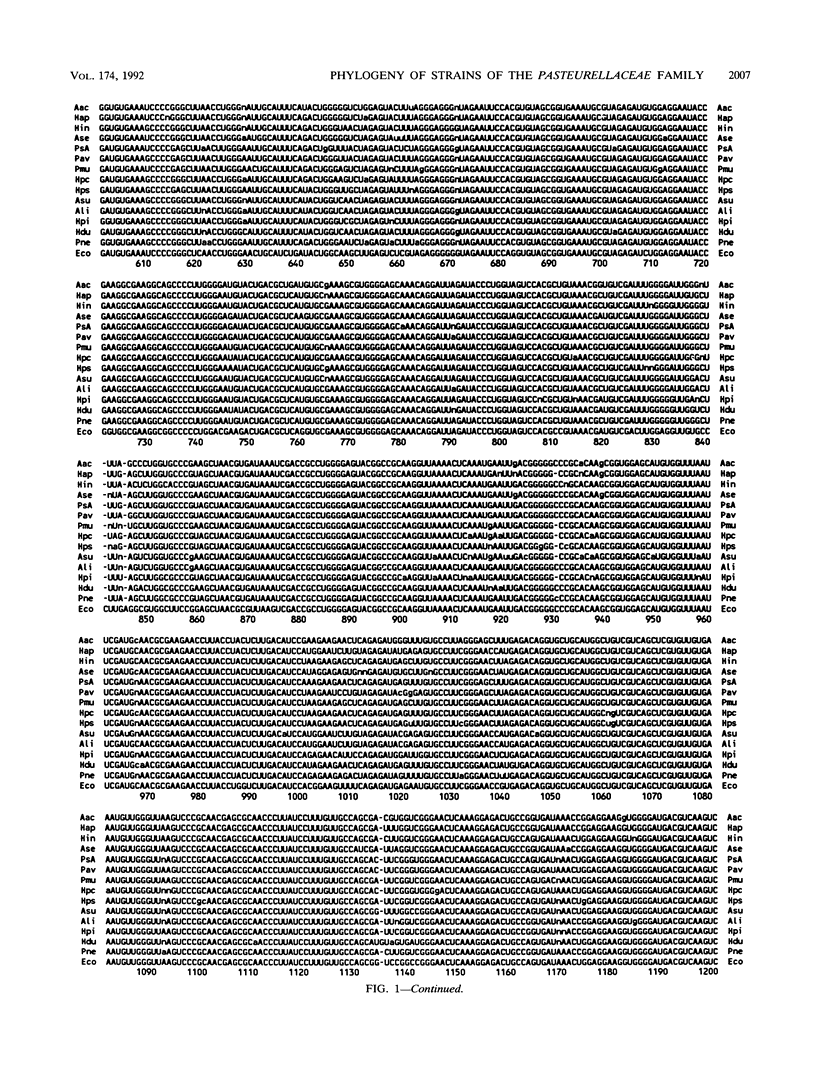
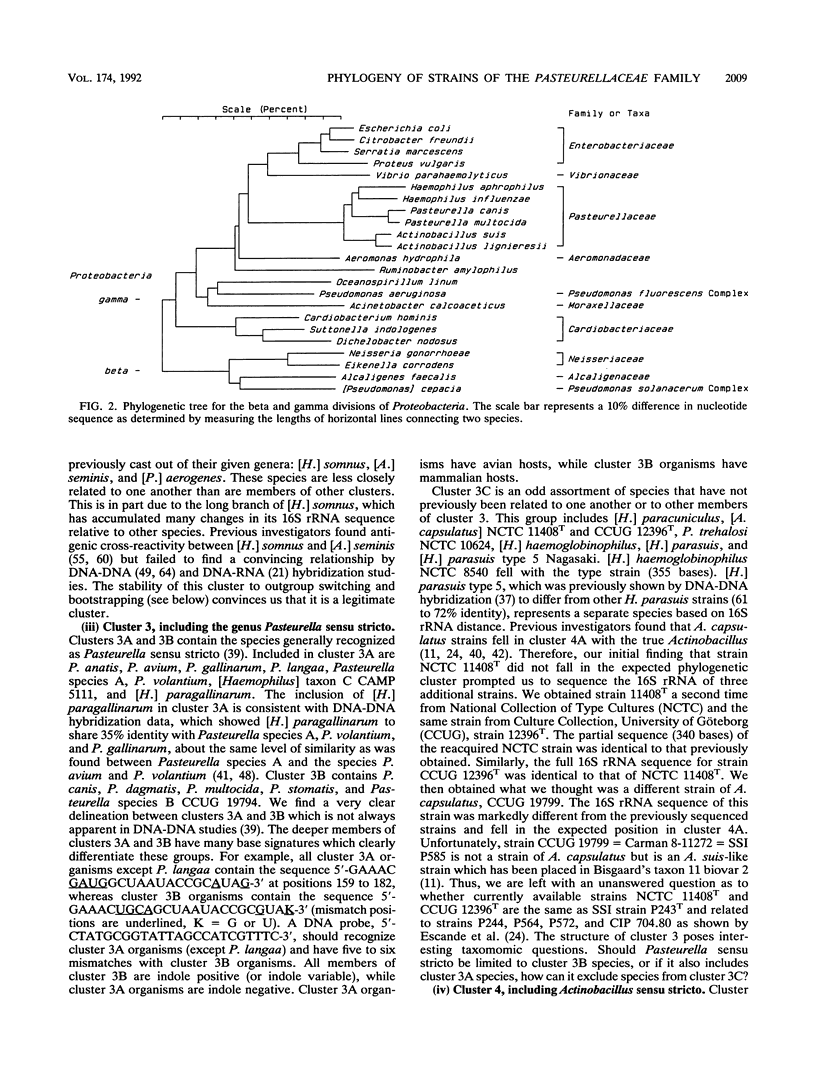
Virtually complete 16S rRNA sequences were determined for 54 representative strains of species in the family Pasteurellaceae. Of these strains, 15 were Pasteurella, 16 were Actinobacillus, and 23 were Haemophilus. A phylogenetic tree was constructed based on sequence similarity, using the Neighbor-Joining method. Fifty-three of the strains fell within four large clusters. The first cluster included the type strains of Haemophilus influenzae, H. aegyptius, H. aphrophilus, H. haemolyticus, H. paraphrophilus, H. segnis, and Actinobacillus actinomycetemcomitans. This cluster also contained A. actinomycetemcomitans FDC Y4, ATCC 29522, ATCC 29523, and ATCC 29524 and H. aphrophilus NCTC 7901. The second cluster included the type strains of A. seminis and Pasteurella aerogenes and H. somnus OVCG 43826. The third cluster was composed of the type strains of Pasteurella multocida, P. anatis, P. avium, P. canis, P. dagmatis, P. gallinarum, P. langaa, P. stomatis, P. volantium, H. haemoglobinophilus, H. parasuis, H. paracuniculus, H. paragallinarum, and A. capsulatus. This cluster also contained Pasteurella species A CCUG 18782, Pasteurella species B CCUG 19974, Haemophilus taxon C CAPM 5111, H. parasuis type 5 Nagasaki, P. volantium (H. parainfluenzae) NCTC 4101, and P. trehalosi NCTC 10624. The fourth cluster included the type strains of Actinobacillus lignieresii, A. equuli, A. pleuropneumoniae, A. suis, A. ureae, H. parahaemolyticus, H. parainfluenzae, H. paraphrohaemolyticus, H. ducreyi, and P. haemolytica. This cluster also contained Actinobacillus species strain CCUG 19799 (Bisgaard taxon 11), A. suis ATCC 15557, H. ducreyi ATCC 27722 and HD 35000, Haemophilus minor group strain 202, and H. parainfluenzae ATCC 29242. The type strain of P. pneumotropica branched alone to form a fifth group. The branching of the Pasteurellaceae family tree was quite complex. The four major clusters contained multiple subclusters. The clusters contained both rapidly and slowly evolving strains (indicated by differing numbers of base changes incorporated into the 16S rRNA sequence relative to outgroup organisms). While the results presented a clear picture of the phylogenetic relationships, the complexity of the branching will make division of the family into genera a difficult and somewhat subjective task. We do not suggest any taxonomic changes at this time.
Full text
PDF








Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Albritton W. L. Biology of Haemophilus ducreyi. Microbiol Rev. 1989 Dec;53(4):377–389. doi: 10.1128/mr.53.4.377-389.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker L. E. Organelle pathology in metabolic neuromuscular disease: an overview. Can J Vet Res. 1990 Jan;54(1):1–14. [PMC free article] [PubMed] [Google Scholar]
- Bisgaard M. Actinobacillus muris sp. nov. isolated from mice. Acta Pathol Microbiol Immunol Scand B. 1986 Feb;94(1):1–8. doi: 10.1111/j.1699-0463.1986.tb03013.x. [DOI] [PubMed] [Google Scholar]
- Bisgaard M. Comparative investigations of Pasteurella haemolytica sensu stricto and so-called P. haemolytica isolated from different pathological lesions in pigs. Acta Pathol Microbiol Immunol Scand B. 1984 Aug;92(4):201–207. doi: 10.1111/j.1699-0463.1984.tb02821.x. [DOI] [PubMed] [Google Scholar]
- Bisgaard M., Mutters R. Characterization of some previously unclassified "Pasteurella" spp. obtained from the oral cavity of dogs and cats and description of a new species tentatively classified with the family Pasteurellaceae Pohl 1981 and provisionally called taxon 16. Acta Pathol Microbiol Immunol Scand B. 1986 Jun;94(3):177–184. doi: 10.1111/j.1699-0463.1986.tb03039.x. [DOI] [PubMed] [Google Scholar]
- Bisgaard M., Mutters R. Re-investigations of selected bovine and ovine strains previously classified as Pasteurella haemolytica and description of some new taxa within the Pasteurella haemolytica-complex. Acta Pathol Microbiol Immunol Scand B. 1986 Jun;94(3):185–193. doi: 10.1111/j.1699-0463.1986.tb03040.x. [DOI] [PubMed] [Google Scholar]
- Bisgaard M., Phillips J. E., Mannheim W. Characterization and identification of bovine and ovine Pasteurellaceae isolated from the oral cavity and rumen of apparently normal cattle and sheep. Acta Pathol Microbiol Immunol Scand B. 1986 Feb;94(1):9–17. doi: 10.1111/j.1699-0463.1986.tb03014.x. [DOI] [PubMed] [Google Scholar]
- Bisgaard M., Piechulla K., Ying Y. T., Frederiksen W., Mannheim W. Prevalence of organisms described as Actinobacillus suis or haemolytic Actinobacillus equuli in the oral cavity of horses. Comparative investigations of strains obtained and porcine strains of A. suis sensu stricto. Acta Pathol Microbiol Immunol Scand B. 1984 Dec;92(6):291–298. doi: 10.1111/j.1699-0463.1984.tb02836.x. [DOI] [PubMed] [Google Scholar]
- Broom A., Sneath P. H. Numerical taxonomy of Haemophilus. J Gen Microbiol. 1981 Sep;126(1):123–149. doi: 10.1099/00221287-126-1-123. [DOI] [PubMed] [Google Scholar]
- Brosius J., Palmer M. L., Kennedy P. J., Noller H. F. Complete nucleotide sequence of a 16S ribosomal RNA gene from Escherichia coli. Proc Natl Acad Sci U S A. 1978 Oct;75(10):4801–4805. doi: 10.1073/pnas.75.10.4801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbon P., Ebel J. P., Ehresmann C. The sequence of the ribosomal 16S RNA from Proteus vulgaris. Sequence comparison with E. coli 16S RNA and its use in secondary model building. Nucleic Acids Res. 1981 May 25;9(10):2325–2333. doi: 10.1093/nar/9.10.2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuba P. J., Bock R., Graf G., Adam T., Göbel U. Comparison of 16S rRNA sequences from the family Pasteurellaceae: phylogenetic relatedness by cluster analysis. J Gen Microbiol. 1988 Jul;134(7):1923–1930. doi: 10.1099/00221287-134-7-1923. [DOI] [PubMed] [Google Scholar]
- De Ley J., Mannheim W., Mutters R., Piechulla K., Tytgat R., Segers P., Bisgaard M., Frederiksen W., Hinz K. H., Vanhoucke M. Inter- and intrafamilial similarities of rRNA cistrons of the Pasteurellaceae. Int J Syst Bacteriol. 1990 Apr;40(2):126–137. doi: 10.1099/00207713-40-2-126. [DOI] [PubMed] [Google Scholar]
- Dewhirst F. E., Paster B. J., La Fontaine S., Rood J. I. Transfer of Kingella indologenes (Snell and Lapage 1976) to the genus Suttonella gen. nov. as Suttonella indologenes comb. nov.; transfer of Bacteroides nodosus (Beveridge 1941) to the genus Dichelobacter gen. nov. as Dichelobacter nodosus comb. nov.; and assignment of the genera Cardiobacterium, Dichelobacter, and Suttonella to Cardiobacteriaceae fam. nov. in the gamma division of Proteobacteria on the basis of 16S rRNA sequence comparisons. Int J Syst Bacteriol. 1990 Oct;40(4):426–433. doi: 10.1099/00207713-40-4-426. [DOI] [PubMed] [Google Scholar]
- Gonzalez I. L., Sylvester J. E., Smith T. F., Stambolian D., Schmickel R. D. Ribosomal RNA gene sequences and hominoid phylogeny. Mol Biol Evol. 1990 May;7(3):203–219. doi: 10.1093/oxfordjournals.molbev.a040600. [DOI] [PubMed] [Google Scholar]
- Kilian M. A taxonomic study of the genus Haemophilus, with the proposal of a new species. J Gen Microbiol. 1976 Mar;93(1):9–62. doi: 10.1099/00221287-93-1-9. [DOI] [PubMed] [Google Scholar]
- Lane D. J., Pace B., Olsen G. J., Stahl D. A., Sogin M. L., Pace N. R. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc Natl Acad Sci U S A. 1985 Oct;82(20):6955–6959. doi: 10.1073/pnas.82.20.6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutters R., Bisgaard M., Pohl S. Taxonomic relationship of selected biogroups of Pasteurella haemolytica as revealed by DNA:DNA hybridizations. Acta Pathol Microbiol Immunol Scand B. 1986 Jun;94(3):195–202. doi: 10.1111/j.1699-0463.1986.tb03041.x. [DOI] [PubMed] [Google Scholar]
- Pace B., Matthews E. A., Johnson K. D., Cantor C. R., Pace N. R. Conserved 5S rRNA complement to tRNA is not required for protein synthesis. Proc Natl Acad Sci U S A. 1982 Jan;79(1):36–40. doi: 10.1073/pnas.79.1.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piechulla K., Hinz K. H., Mannheim W. Genetic and phenotypic comparison of three new avian Haemophilus-like taxa and of Haemophilus paragallinarum Biberstein and White 1969 with other members of the family Pasteurellaceae Pohl 1981. Avian Dis. 1985 Jul-Sep;29(3):601–612. [PubMed] [Google Scholar]
- Rahaley R. S. Serological comparison between Histophilus ovis, Actinobacillus seminis and Brucella ovis. Aust Vet J. 1978 Sep;54(9):423–425. doi: 10.1111/j.1751-0813.1978.tb05567.x. [DOI] [PubMed] [Google Scholar]
- Rossau R., Duhamel M., Jannes G., Decourt J. L., Van Heuverswyn H. The development of specific rRNA-derived oligonucleotide probes for Haemophilus ducreyi, the causative agent of chancroid. J Gen Microbiol. 1991 Feb;137(2):277–285. doi: 10.1099/00221287-137-2-277. [DOI] [PubMed] [Google Scholar]
- Rossau R., Heyndrickx L., Van Heuverswyn H. Nucleotide sequence of a 16S ribosomal RNA gene from Neisseria gonorrhoeae. Nucleic Acids Res. 1988 Jul 11;16(13):6227–6227. doi: 10.1093/nar/16.13.6227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987 Jul;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Sneath P. H., Stevens M. A numerical taxonomic study of Actinobacillus, Pasteurella and Yersinia. J Gen Microbiol. 1985 Oct;131(10):2711–2738. doi: 10.1099/00221287-131-10-2711. [DOI] [PubMed] [Google Scholar]
- Stephens L. R., Humphrey J. D., Little P. B., Barnum D. A. Morphological, biochemical, antigenic, and cytochemical relationships among Haemophilus somnus, Haemophilus agni, Haemophilus haemoglobinophilus, Histophilus ovis, and Actinobacillus seminis. J Clin Microbiol. 1983 May;17(5):728–737. doi: 10.1128/jcm.17.5.728-737.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier J. A., Keppler K. J. A note on the neighbor-joining algorithm of Saitou and Nei. Mol Biol Evol. 1988 Nov;5(6):729–731. doi: 10.1093/oxfordjournals.molbev.a040527. [DOI] [PubMed] [Google Scholar]
- Tanner A. C., Visconti R. A., Socransky S. S., Holt S. C. Classification and identification of Actinobacillus actinomycetemcomitans and haemophilus aphrophilus by cluster analysis and deoxyribonucleic acid hybridizations. J Periodontal Res. 1982 Nov;17(6):585–596. doi: 10.1111/j.1600-0765.1982.tb01180.x. [DOI] [PubMed] [Google Scholar]
- Winslow C. E., Broadhurst J., Buchanan R. E., Krumwiede C., Rogers L. A., Smith G. H. The Families and Genera of the Bacteria: Preliminary Report of the Committee of the Society of American Bacteriologists on Characterization and Classification of Bacterial Types. J Bacteriol. 1917 Sep;2(5):505–566. doi: 10.1128/jb.2.5.505-566.1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R. Bacterial evolution. Microbiol Rev. 1987 Jun;51(2):221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R., Stackebrandt E., Macke T. J., Fox G. E. A phylogenetic definition of the major eubacterial taxa. Syst Appl Microbiol. 1985;6:143–151. doi: 10.1016/s0723-2020(85)80047-3. [DOI] [PubMed] [Google Scholar]
- Zambon J. J., Slots J., Genco R. J. Serology of oral Actinobacillus actinomycetemcomitans and serotype distribution in human periodontal disease. Infect Immun. 1983 Jul;41(1):19–27. doi: 10.1128/iai.41.1.19-27.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]


