Abstract
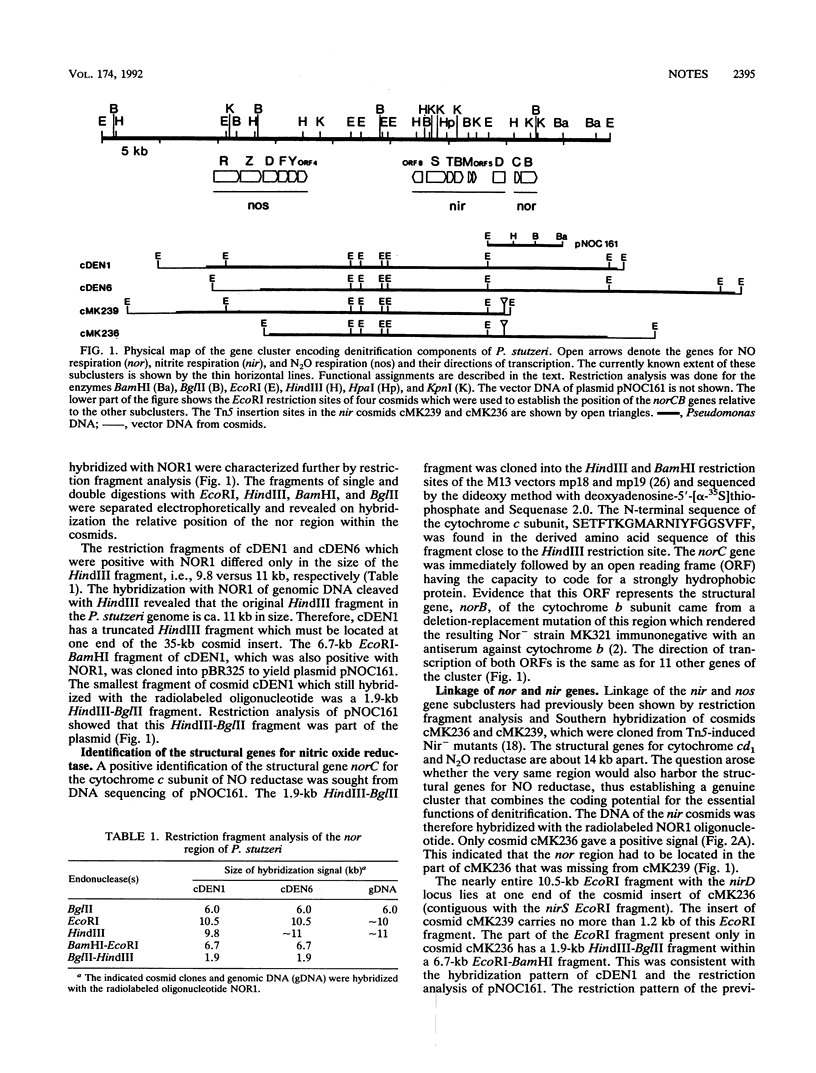
A gene cluster of 30 kilobases required for denitrification in Pseudomonas stutzeri ZoBell was identified and mapped. It harbors genes necessary for the respiratory reduction of nitrite (nir genes), nitric oxide (nor genes), and nitrous oxide (nos genes). Fifteen genes, 13 of which are transcribed in the same direction, have been located on a 56-kb BamHI fragment. They are arranged in three subclusters in the order nos-nir-nor.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Boyer H. W., Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969 May 14;41(3):459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- Braun C., Zumft W. G. Marker exchange of the structural genes for nitric oxide reductase blocks the denitrification pathway of Pseudomonas stutzeri at nitric oxide. J Biol Chem. 1991 Dec 5;266(34):22785–22788. [PubMed] [Google Scholar]
- Carr G. J., Ferguson S. J. The nitric oxide reductase of Paracoccus denitrificans. Biochem J. 1990 Jul 15;269(2):423–429. doi: 10.1042/bj2690423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr G. J., Page M. D., Ferguson S. J. The energy-conserving nitric-oxide-reductase system in Paracoccus denitrificans. Distinction from the nitrite reductase that catalyses synthesis of nitric oxide and evidence from trapping experiments for nitric oxide as a free intermediate during denitrification. Eur J Biochem. 1989 Feb 15;179(3):683–692. doi: 10.1111/j.1432-1033.1989.tb14601.x. [DOI] [PubMed] [Google Scholar]
- Chu G., Vollrath D., Davis R. W. Separation of large DNA molecules by contour-clamped homogeneous electric fields. Science. 1986 Dec 19;234(4783):1582–1585. doi: 10.1126/science.3538420. [DOI] [PubMed] [Google Scholar]
- Grunstein M., Hogness D. S. Colony hybridization: a method for the isolation of cloned DNAs that contain a specific gene. Proc Natl Acad Sci U S A. 1975 Oct;72(10):3961–3965. doi: 10.1073/pnas.72.10.3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heery D. M., Gannon F., Powell R. A simple method for subcloning DNA fragments from gel slices. Trends Genet. 1990 Jun;6(6):173–173. doi: 10.1016/0168-9525(90)90158-3. [DOI] [PubMed] [Google Scholar]
- Heiss B., Frunzke K., Zumft W. G. Formation of the N-N bond from nitric oxide by a membrane-bound cytochrome bc complex of nitrate-respiring (denitrifying) Pseudomonas stutzeri. J Bacteriol. 1989 Jun;171(6):3288–3297. doi: 10.1128/jb.171.6.3288-3297.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochstein L. I., Tomlinson G. A. The enzymes associated with denitrification. Annu Rev Microbiol. 1988;42:231–261. doi: 10.1146/annurev.mi.42.100188.001311. [DOI] [PubMed] [Google Scholar]
- Holloway B. W., Morgan A. F. Genome organization in Pseudomonas. Annu Rev Microbiol. 1986;40:79–105. doi: 10.1146/annurev.mi.40.100186.000455. [DOI] [PubMed] [Google Scholar]
- Jackson M. A., Tiedje J. M., Averill B. A. Evidence for a NO-rebound mechanism for production of N2O from nitrite by the copper-containing nitrite reductase from Achromobacter cycloclastes. FEBS Lett. 1991 Oct 7;291(1):41–44. doi: 10.1016/0014-5793(91)81099-t. [DOI] [PubMed] [Google Scholar]
- Jüngst A., Braun C., Zumft W. G. Close linkage in Pseudomonas stutzeri of the structural genes for respiratory nitrite reductase and nitrous oxide reductase, and other essential genes for denitrification. Mol Gen Genet. 1991 Feb;225(2):241–248. doi: 10.1007/BF00269855. [DOI] [PubMed] [Google Scholar]
- Jüngst A., Wakabayashi S., Matsubara H., Zumft W. G. The nirSTBM region coding for cytochrome cd1-dependent nitrite respiration of Pseudomonas stutzeri consists of a cluster of mono-, di-, and tetraheme proteins. FEBS Lett. 1991 Feb 25;279(2):205–209. doi: 10.1016/0014-5793(91)80150-2. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Leary J. J., Brigati D. J., Ward D. C. Rapid and sensitive colorimetric method for visualizing biotin-labeled DNA probes hybridized to DNA or RNA immobilized on nitrocellulose: Bio-blots. Proc Natl Acad Sci U S A. 1983 Jul;80(13):4045–4049. doi: 10.1073/pnas.80.13.4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H. S., Abdelal A. H., Clark M. A., Ingraham J. L. Molecular characterization of nosA, a Pseudomonas stutzeri gene encoding an outer membrane protein required to make copper-containing N2O reductase. J Bacteriol. 1991 Sep;173(17):5406–5413. doi: 10.1128/jb.173.17.5406-5413.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Römermann D., Friedrich B. Denitrification by Alcaligenes eutrophus is plasmid dependent. J Bacteriol. 1985 May;162(2):852–854. doi: 10.1128/jb.162.2.852-854.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stouthamer A. H. Metabolic regulation including anaerobic metabolism in Paracoccus denitrificans. J Bioenerg Biomembr. 1991 Apr;23(2):163–185. doi: 10.1007/BF00762216. [DOI] [PubMed] [Google Scholar]
- Ye R. W., Toro-Suarez I., Tiedje J. M., Averill B. A. H218O isotope exchange studies on the mechanism of reduction of nitric oxide and nitrite to nitrous oxide by denitrifying bacteria. Evidence for an electrophilic nitrosyl during reduction of nitric oxide. J Biol Chem. 1991 Jul 15;266(20):12848–12851. [PubMed] [Google Scholar]
- Zumft W. G., Döhler K., Körner H., Löchelt S., Viebrock A., Frunzke K. Defects in cytochrome cd1-dependent nitrite respiration of transposon Tn5-induced mutants from Pseudomonas stutzeri. Arch Microbiol. 1988;149(6):492–498. doi: 10.1007/BF00446750. [DOI] [PubMed] [Google Scholar]
- Zumft W. G., Viebrock-Sambale A., Braun C. Nitrous oxide reductase from denitrifying Pseudomonas stutzeri. Genes for copper-processing and properties of the deduced products, including a new member of the family of ATP/GTP-binding proteins. Eur J Biochem. 1990 Sep 24;192(3):591–599. doi: 10.1111/j.1432-1033.1990.tb19265.x. [DOI] [PubMed] [Google Scholar]