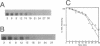Abstract
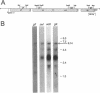
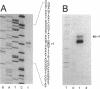
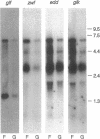
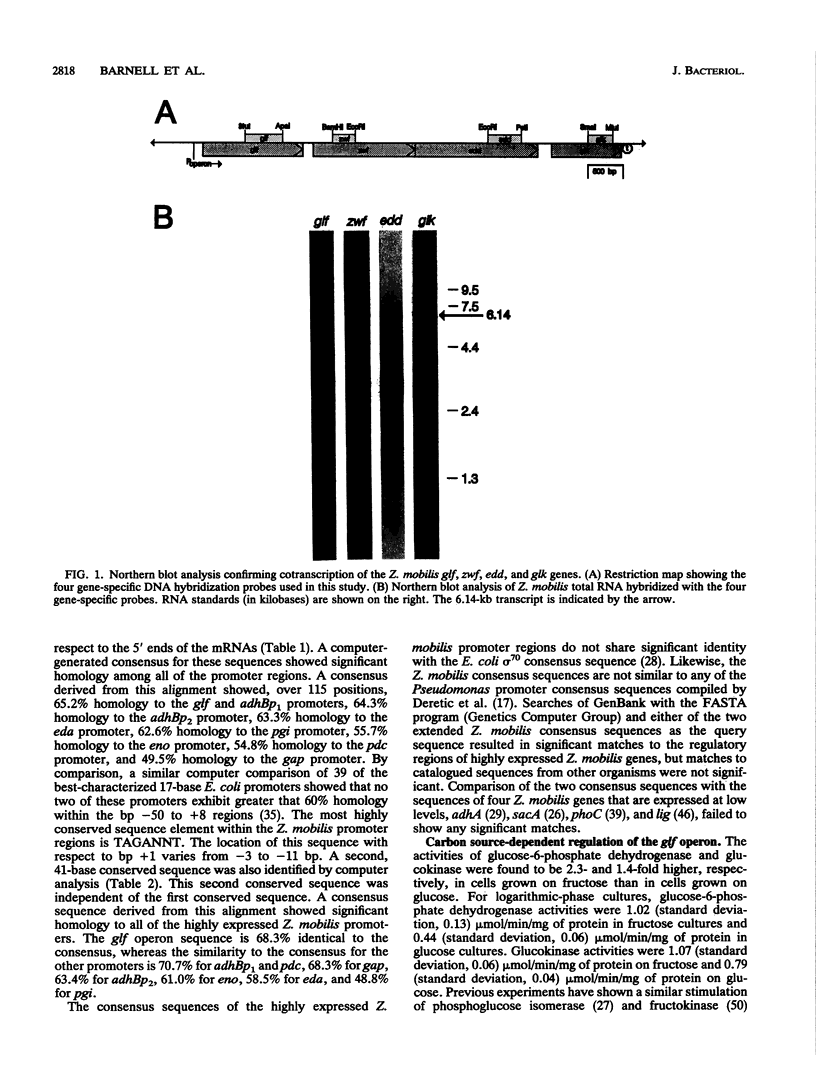
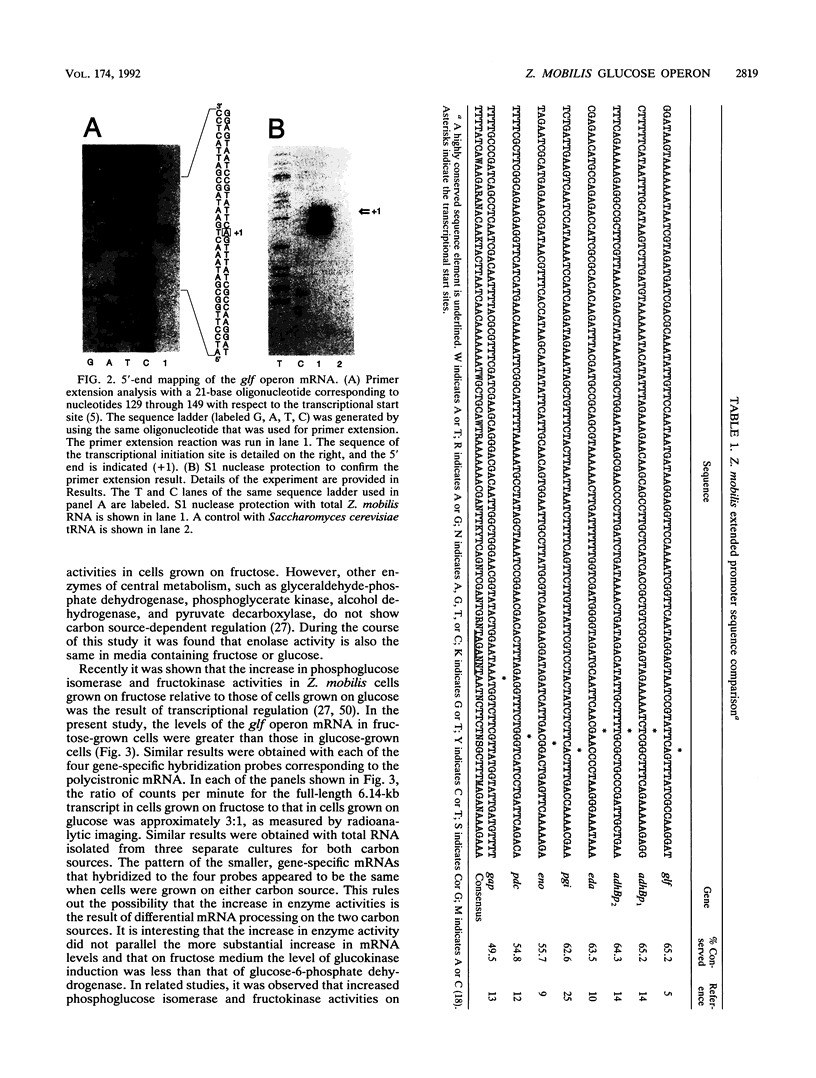
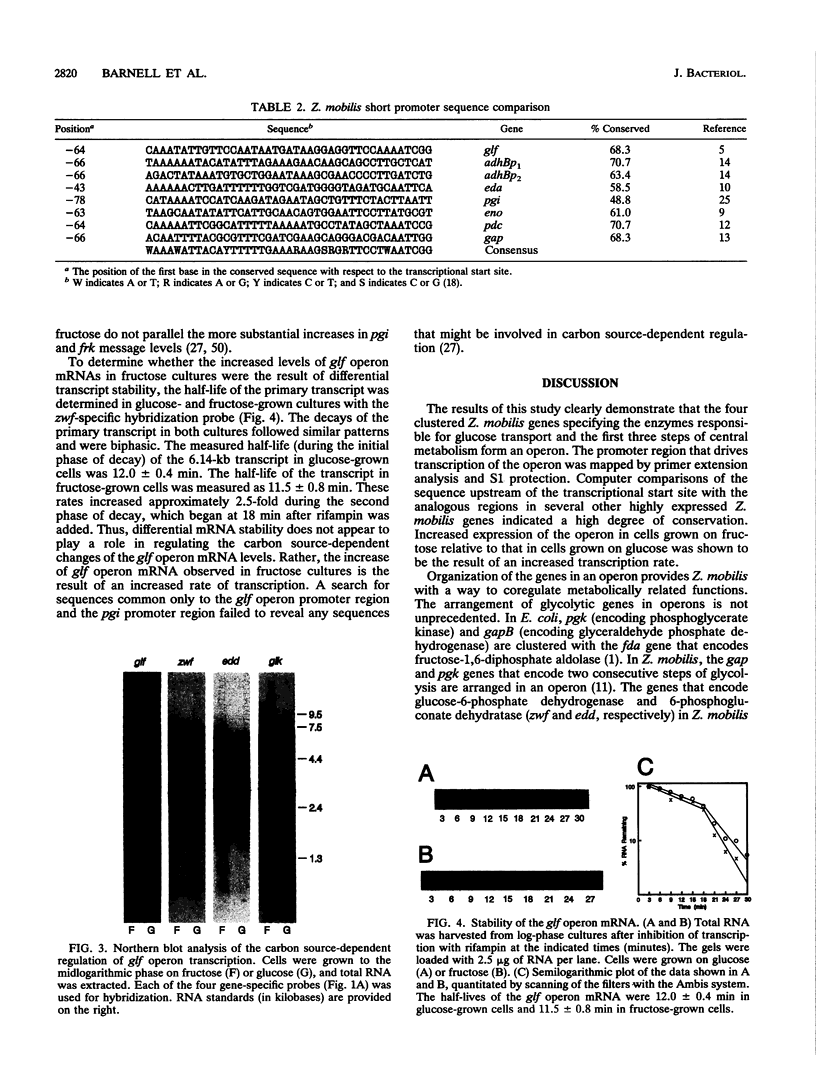
The Zymomonas mobilis genes that encode the glucose-facilitated diffusion transporter (glf), glucose-6-phosphate dehydrogenase (zwf), 6-phosphogluconate dehydratase (edd), and glucokinase (glk) are clustered on the genome. The data presented here firmly establish that the glf, zwf, edd, and glk genes form an operon, in that order. The four genes of the operon are cotranscribed on a 6.14-kb mRNA. The site of transcriptional initiation for the polycistronic message was mapped by primer extension and nuclease S1 protection analysis. The glf operon promoter region showed significant homology to other highly expressed Z. mobilis promoters, but not to consensus promoters from other bacteria. The highly expressed Z. mobilis promoter set contains two independent, overlapping, conserved sequences that extend from approximately bp -100 to +15 with respect to the transcriptional start sites. Expression of the glf operon was shown to be subject to carbon source-dependent regulation. The mRNA level was threefold higher in cells grown on fructose than in cells grown on glucose. This increase was not the result of differential mRNA processing when cells were grown on the different carbon sources, nor was it the result of differential transcript stability. Degradation of the 6.14-kb glf operon mRNA was biphasic, with initial half-lives of 11.5 min in fructose-grown cells and 12.0 min in glucose-grown cells. Thus, the higher level of glf operon mRNA in fructose-grown cells is the result of an increased rate of transcription. The importance of increasing glf expression in cells growing on fructose is discussed.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alefounder P. R., Perham R. N. Identification, molecular cloning and sequence analysis of a gene cluster encoding the class II fructose 1,6-bisphosphate aldolase, 3-phosphoglycerate kinase and a putative second glyceraldehyde 3-phosphate dehydrogenase of Escherichia coli. Mol Microbiol. 1989 Jun;3(6):723–732. doi: 10.1111/j.1365-2958.1989.tb00221.x. [DOI] [PubMed] [Google Scholar]
- An H., Scopes R. K., Rodriguez M., Keshav K. F., Ingram L. O. Gel electrophoretic analysis of Zymomonas mobilis glycolytic and fermentative enzymes: identification of alcohol dehydrogenase II as a stress protein. J Bacteriol. 1991 Oct;173(19):5975–5982. doi: 10.1128/jb.173.19.5975-5982.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann B. J. Linkage map of Escherichia coli K-12, edition 8. Microbiol Rev. 1990 Jun;54(2):130–197. doi: 10.1128/mr.54.2.130-197.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnell W. O., Yi K. C., Conway T. Sequence and genetic organization of a Zymomonas mobilis gene cluster that encodes several enzymes of glucose metabolism. J Bacteriol. 1990 Dec;172(12):7227–7240. doi: 10.1128/jb.172.12.7227-7240.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisson L. F., Fraenkel D. G. Expression of kinase-dependent glucose uptake in Saccharomyces cerevisiae. J Bacteriol. 1984 Sep;159(3):1013–1017. doi: 10.1128/jb.159.3.1013-1017.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway T., Fliege R., Jones-Kilpatrick D., Liu J., Barnell W. O., Egan S. E. Cloning, characterization and expression of the Zymononas mobilis eda gene that encodes 2-keto-3-deoxy-6-phosphogluconate aldolase of the Entner-Doudoroff pathway. Mol Microbiol. 1991 Dec;5(12):2901–2911. doi: 10.1111/j.1365-2958.1991.tb01850.x. [DOI] [PubMed] [Google Scholar]
- Conway T., Ingram L. O. Phosphoglycerate kinase gene from Zymomonas mobilis: cloning, sequencing, and localization within the gap operon. J Bacteriol. 1988 Apr;170(4):1926–1933. doi: 10.1128/jb.170.4.1926-1933.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway T., Ingram L. O. Similarity of Escherichia coli propanediol oxidoreductase (fucO product) and an unusual alcohol dehydrogenase from Zymomonas mobilis and Saccharomyces cerevisiae. J Bacteriol. 1989 Jul;171(7):3754–3759. doi: 10.1128/jb.171.7.3754-3759.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway T., Osman Y. A., Konnan J. I., Hoffmann E. M., Ingram L. O. Promoter and nucleotide sequences of the Zymomonas mobilis pyruvate decarboxylase. J Bacteriol. 1987 Mar;169(3):949–954. doi: 10.1128/jb.169.3.949-954.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway T., Sewell G. W., Ingram L. O. Glyceraldehyde-3-phosphate dehydrogenase gene from Zymomonas mobilis: cloning, sequencing, and identification of promoter region. J Bacteriol. 1987 Dec;169(12):5653–5662. doi: 10.1128/jb.169.12.5653-5662.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway T., Sewell G. W., Osman Y. A., Ingram L. O. Cloning and sequencing of the alcohol dehydrogenase II gene from Zymomonas mobilis. J Bacteriol. 1987 Jun;169(6):2591–2597. doi: 10.1128/jb.169.6.2591-2597.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway T., Yi K. C., Egan S. E., Wolf R. E., Jr, Rowley D. L. Locations of the zwf, edd, and eda genes on the Escherichia coli physical map. J Bacteriol. 1991 Sep;173(17):5247–5248. doi: 10.1128/jb.173.17.5247-5248.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimarco A. A., Romano A. H. d-Glucose Transport System of Zymomonas mobilis. Appl Environ Microbiol. 1985 Jan;49(1):151–157. doi: 10.1128/aem.49.1.151-157.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy C. K., Mejia J. P., Conway T., Ingram L. O. Differential expression of gap and pgk genes within the gap operon of Zymomonas mobilis. J Bacteriol. 1989 Dec;171(12):6549–6554. doi: 10.1128/jb.171.12.6549-6554.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesman T. L., Barnell W. O., Conway T. Cloning, characterization, and nucleotide sequence analysis of a Zymomonas mobilis phosphoglucose isomerase gene that is subject to carbon source-dependent regulation. J Bacteriol. 1991 May;173(10):3215–3223. doi: 10.1128/jb.173.10.3215-3223.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keshav K. F., Yomano L. P., An H. J., Ingram L. O. Cloning of the Zymomonas mobilis structural gene encoding alcohol dehydrogenase I (adhA): sequence comparison and expression in Escherichia coli. J Bacteriol. 1990 May;172(5):2491–2497. doi: 10.1128/jb.172.5.2491-2497.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Barnell W. O., Conway T. The polycistronic mRNA of the Zymomonas mobilis glf-zwf-edd-glk operon is subject to complex transcript processing. J Bacteriol. 1992 May;174(9):2824–2833. doi: 10.1128/jb.174.9.2824-2833.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luria S. E., Delbrück M. Mutations of Bacteria from Virus Sensitivity to Virus Resistance. Genetics. 1943 Nov;28(6):491–511. doi: 10.1093/genetics/28.6.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie K. F., Conway T., Aldrich H. C., Ingram L. O. Expression of Zymomonas mobilis adhB (encoding alcohol dehydrogenase II) and adhB-lacZ operon fusions in recombinant Z. mobilis. J Bacteriol. 1989 Sep;171(9):4577–4582. doi: 10.1128/jb.171.9.4577-4582.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackie G. A. Stabilization of the 3' one-third of Escherichia coli ribosomal protein S20 mRNA in mutants lacking polynucleotide phosphorylase. J Bacteriol. 1989 Aug;171(8):4112–4120. doi: 10.1128/jb.171.8.4112-4120.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill M. C., Chiafari F. Escherichia coli promoters. II. A spacing class-dependent promoter search protocol. J Biol Chem. 1989 Apr 5;264(10):5531–5534. [PubMed] [Google Scholar]
- O'Neill M. C. Escherichia coli promoters. I. Consensus as it relates to spacing class, specificity, repeat substructure, and three-dimensional organization. J Biol Chem. 1989 Apr 5;264(10):5522–5530. [PubMed] [Google Scholar]
- Osman Y. A., Conway T., Bonetti S. J., Ingram L. O. Glycolytic flux in Zymomonas mobilis: enzyme and metabolite levels during batch fermentation. J Bacteriol. 1987 Aug;169(8):3726–3736. doi: 10.1128/jb.169.8.3726-3736.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pond J. L., Eddy C. K., Mackenzie K. F., Conway T., Borecky D. J., Ingram L. O. Cloning, sequencing, and characterization of the principal acid phosphatase, the phoC+ product, from Zymomonas mobilis. J Bacteriol. 1989 Feb;171(2):767–774. doi: 10.1128/jb.171.2.767-774.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowley D. L., Wolf R. E., Jr Molecular characterization of the Escherichia coli K-12 zwf gene encoding glucose 6-phosphate dehydrogenase. J Bacteriol. 1991 Feb;173(3):968–977. doi: 10.1128/jb.173.3.968-977.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scopes R. K., Griffiths-Smith K. Use of differential dye-ligand chromatography with affinity elution for enzyme purification: 6-phosphogluconate dehydratase from Zymomonas mobilis. Anal Biochem. 1984 Feb;136(2):530–534. doi: 10.1016/0003-2697(84)90257-4. [DOI] [PubMed] [Google Scholar]
- Scopes R. K., Testolin V., Stoter A., Griffiths-Smith K., Algar E. M. Simultaneous purification and characterization of glucokinase, fructokinase and glucose-6-phosphate dehydrogenase from Zymomonas mobilis. Biochem J. 1985 Jun 15;228(3):627–634. doi: 10.1042/bj2280627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temple L., Cuskey S. M., Perkins R. E., Bass R. C., Morales N. M., Christie G. E., Olsen R. H., Phibbs P. V., Jr Analysis of cloned structural and regulatory genes for carbohydrate utilization in Pseudomonas aeruginosa PAO. J Bacteriol. 1990 Nov;172(11):6396–6402. doi: 10.1128/jb.172.11.6396-6402.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanBogelen R. A., Hutton M. E., Neidhardt F. C. Gene-protein database of Escherichia coli K-12: edition 3. Electrophoresis. 1990 Dec;11(12):1131–1166. doi: 10.1002/elps.1150111205. [DOI] [PubMed] [Google Scholar]