Abstract
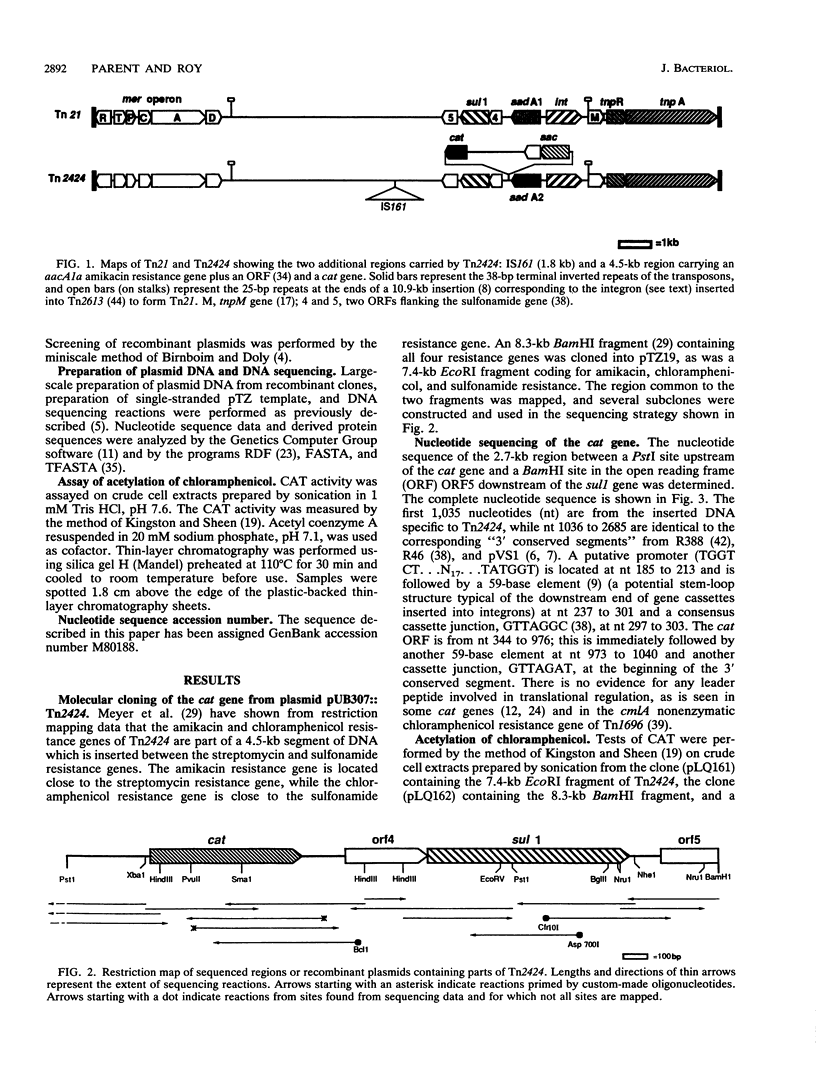
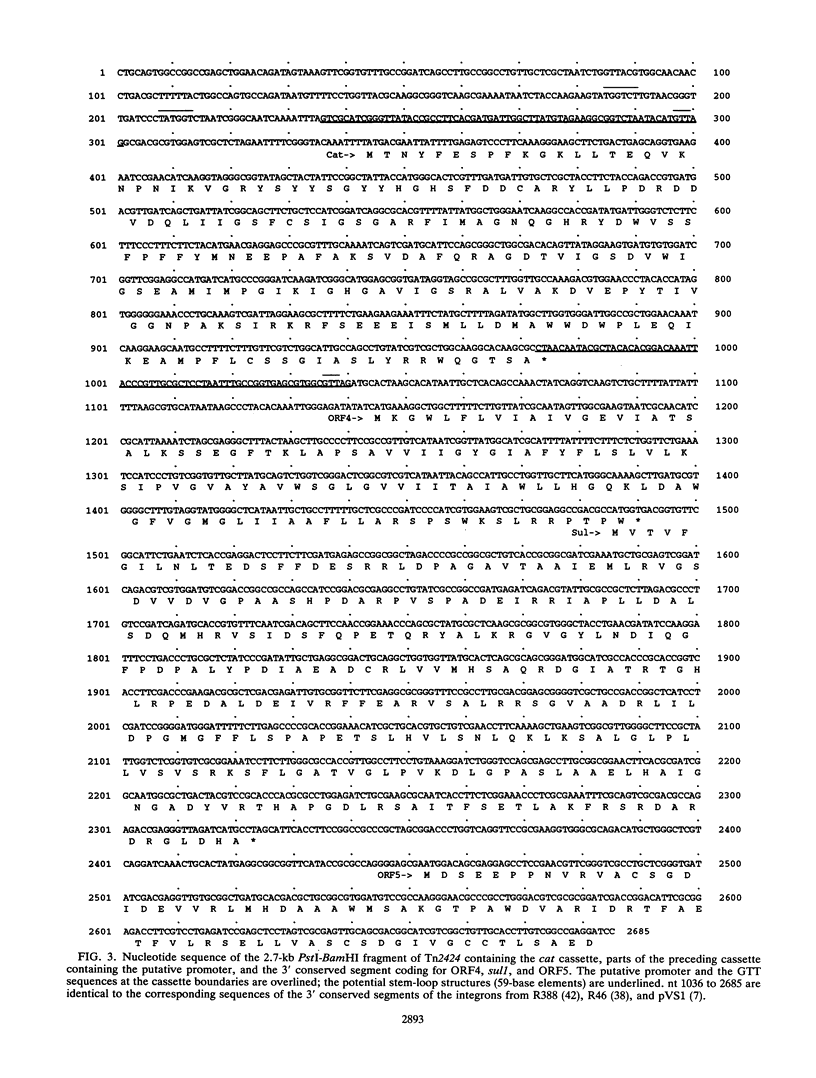
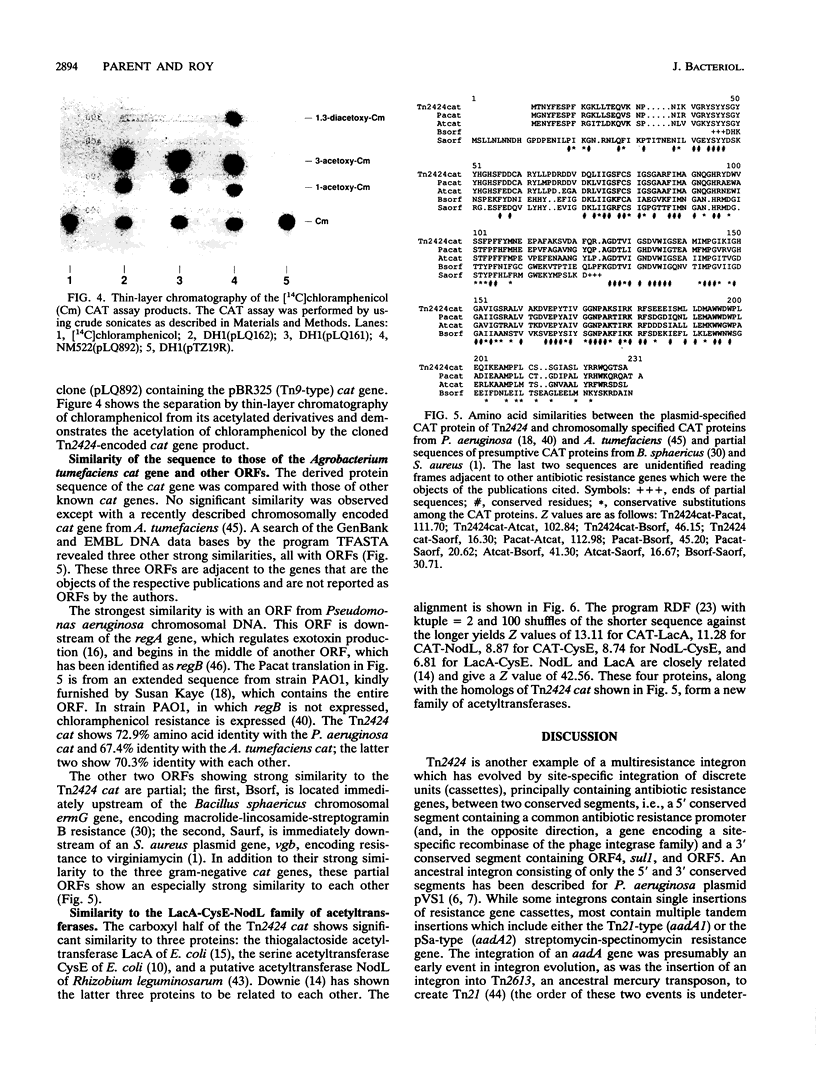
We have sequenced the gene coding for the chloramphenicol acetyltransferase of Tn2424 of plasmid NR79. This gene codes for a protein of 23,500 Da, and the derived protein sequence is similar to those of the chromosomal chloramphenicol acetyltransferases of Agrobacterium tumefaciens and Pseudomonas aeruginosa and of unidentified open reading frames, which may encode chloramphenicol acetyltransferases, adjacent to the ermG macrolide-lincosamide-streptogramin resistance gene of Bacillus sphaericus and the vgb virginiamycin resistance gene of Staphylococcus aureus. Weaker similarity to the LacA (thiogalactoside acetyltransferase) and CysE (serine acetyltransferase) proteins of Escherichia coli and the NodL protein of Rhizobium leguminosarum is also observed. There is no significant similarity to any other chloramphenicol acetyltransferase genes, such as that of Tn9. The Tn2424 cat gene is part of a 4.5-kb region which also contains the aacA1a aminoglycoside-6'-N-acetyltransferase gene; Tn2424 is similar to Tn21 except for the presence of this region. Sequences flanking the cat gene are typical of those flanking other genes inserted into pVS1-derived "integrons" by a site-specific recombinational mechanism.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allignet J., Loncle V., Mazodier P., el Solh N. Nucleotide sequence of a staphylococcal plasmid gene, vgb, encoding a hydrolase inactivating the B components of virginiamycin-like antibiotics. Plasmid. 1988 Nov;20(3):271–275. doi: 10.1016/0147-619x(88)90034-0. [DOI] [PubMed] [Google Scholar]
- Bannam T. L., Rood J. I. Relationship between the Clostridium perfringens catQ gene product and chloramphenicol acetyltransferases from other bacteria. Antimicrob Agents Chemother. 1991 Mar;35(3):471–476. doi: 10.1128/aac.35.3.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaulieu D., Ouellette M., Bergeron M. G., Roy P. H. Characterization of a plasmid isolated from Branhamella catarrhalis and detection of plasmid sequences within the genome of a B. catarrhalis strain. Plasmid. 1988 Sep;20(2):158–162. doi: 10.1016/0147-619x(88)90020-0. [DOI] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bissonnette L., Champetier S., Buisson J. P., Roy P. H. Characterization of the nonenzymatic chloramphenicol resistance (cmlA) gene of the In4 integron of Tn1696: similarity of the product to transmembrane transport proteins. J Bacteriol. 1991 Jul;173(14):4493–4502. doi: 10.1128/jb.173.14.4493-4502.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bissonnette L., Roy P. H. Characterization of In0 of Pseudomonas aeruginosa plasmid pVS1, an ancestor of integrons of multiresistance plasmids and transposons of gram-negative bacteria. J Bacteriol. 1992 Feb;174(4):1248–1257. doi: 10.1128/jb.174.4.1248-1257.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown N. L., Misra T. K., Winnie J. N., Schmidt A., Seiff M., Silver S. The nucleotide sequence of the mercuric resistance operons of plasmid R100 and transposon Tn501: further evidence for mer genes which enhance the activity of the mercuric ion detoxification system. Mol Gen Genet. 1986 Jan;202(1):143–151. doi: 10.1007/BF00330531. [DOI] [PubMed] [Google Scholar]
- Cameron F. H., Groot Obbink D. J., Ackerman V. P., Hall R. M. Nucleotide sequence of the AAD(2'') aminoglycoside adenylyltransferase determinant aadB. Evolutionary relationship of this region with those surrounding aadA in R538-1 and dhfrII in R388. Nucleic Acids Res. 1986 Nov 11;14(21):8625–8635. doi: 10.1093/nar/14.21.8625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denk D., Böck A. L-cysteine biosynthesis in Escherichia coli: nucleotide sequence and expression of the serine acetyltransferase (cysE) gene from the wild-type and a cysteine-excreting mutant. J Gen Microbiol. 1987 Mar;133(3):515–525. doi: 10.1099/00221287-133-3-515. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick T., Matzura H. Chloramphenicol-induced translational activation of cat messenger RNA in vitro. J Mol Biol. 1990 Apr 20;212(4):661–668. doi: 10.1016/0022-2836(90)90228-E. [DOI] [PubMed] [Google Scholar]
- Dorman C. J., Foster T. J. Nonenzymatic chloramphenicol resistance determinants specified by plasmids R26 and R55-1 in Escherichia coli K-12 do not confer high-level resistance to fluorinated analogs. Antimicrob Agents Chemother. 1982 Nov;22(5):912–914. doi: 10.1128/aac.22.5.912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downie J. A. The nodL gene from Rhizobium leguminosarum is homologous to the acetyl transferases encoded by lacA and cysE. Mol Microbiol. 1989 Nov;3(11):1649–1651. doi: 10.1111/j.1365-2958.1989.tb00150.x. [DOI] [PubMed] [Google Scholar]
- Hediger M. A., Johnson D. F., Nierlich D. P., Zabin I. DNA sequence of the lactose operon: the lacA gene and the transcriptional termination region. Proc Natl Acad Sci U S A. 1985 Oct;82(19):6414–6418. doi: 10.1073/pnas.82.19.6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindahl M. S., Frank D. W., Hamood A., Iglewski B. H. Characterization of a gene that regulates toxin A synthesis in Pseudomonas aeruginosa. Nucleic Acids Res. 1988 Jun 24;16(12):5699–5699. doi: 10.1093/nar/16.12.5699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyde D. R., Tu C. P. tnpM: a novel regulatory gene that enhances Tn21 transposition and suppresses cointegrate resolution. Cell. 1985 Sep;42(2):629–638. doi: 10.1016/0092-8674(85)90120-5. [DOI] [PubMed] [Google Scholar]
- Le Grice S. F., Matzura H., Marcoli R., Iida S., Bickle T. A. The catabolite-sensitive promoter for the chloramphenicol acetyl transferase gene is preceded by two binding sites for the catabolite gene activator protein. J Bacteriol. 1982 Apr;150(1):312–318. doi: 10.1128/jb.150.1.312-318.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie A. G., Moody P. C., Shaw W. V. Structure of chloramphenicol acetyltransferase at 1.75-A resolution. Proc Natl Acad Sci U S A. 1988 Jun;85(12):4133–4137. doi: 10.1073/pnas.85.12.4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie A. G. Refined crystal structure of type III chloramphenicol acetyltransferase at 1.75 A resolution. J Mol Biol. 1990 May 5;213(1):167–186. doi: 10.1016/S0022-2836(05)80129-9. [DOI] [PubMed] [Google Scholar]
- Lipman D. J., Pearson W. R. Rapid and sensitive protein similarity searches. Science. 1985 Mar 22;227(4693):1435–1441. doi: 10.1126/science.2983426. [DOI] [PubMed] [Google Scholar]
- Lovett P. S. Translational attenuation as the regulator of inducible cat genes. J Bacteriol. 1990 Jan;172(1):1–6. doi: 10.1128/jb.172.1.1-6.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez E., de la Cruz F. Genetic elements involved in Tn21 site-specific integration, a novel mechanism for the dissemination of antibiotic resistance genes. EMBO J. 1990 Apr;9(4):1275–1281. doi: 10.1002/j.1460-2075.1990.tb08236.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercier J., Lachapelle J., Couture F., Lafond M., Vézina G., Boissinot M., Levesque R. C. Structural and functional characterization of tnpI, a recombinase locus in Tn21 and related beta-lactamase transposons. J Bacteriol. 1990 Jul;172(7):3745–3757. doi: 10.1128/jb.172.7.3745-3757.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer J. F., Nies B. A., Kratz J., Wiedemann B. Evolution of Tn21-related transposons: isolation of Tn2425, which harbours IS161. J Gen Microbiol. 1985 May;131(5):1123–1130. doi: 10.1099/00221287-131-5-1123. [DOI] [PubMed] [Google Scholar]
- Meyer J. F., Nies B. A., Wiedemann B. Amikacin resistance mediated by multiresistance transposon Tn2424. J Bacteriol. 1983 Aug;155(2):755–760. doi: 10.1128/jb.155.2.755-760.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monod M., Mohan S., Dubnau D. Cloning and analysis of ermG, a new macrolide-lincosamide-streptogramin B resistance element from Bacillus sphaericus. J Bacteriol. 1987 Jan;169(1):340–350. doi: 10.1128/jb.169.1.340-350.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouellette M., Bissonnette L., Roy P. H. Precise insertion of antibiotic resistance determinants into Tn21-like transposons: nucleotide sequence of the OXA-1 beta-lactamase gene. Proc Natl Acad Sci U S A. 1987 Nov;84(21):7378–7382. doi: 10.1073/pnas.84.21.7378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouellette M., Roy P. H. Homology of ORFs from Tn2603 and from R46 to site-specific recombinases. Nucleic Acids Res. 1987 Dec 10;15(23):10055–10055. doi: 10.1093/nar/15.23.10055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson W. R., Lipman D. J. Improved tools for biological sequence comparison. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pothier F., Ouellet M., Julien J. P., Guérin S. L. An improved CAT assay for promoter analysis in either transgenic mice or tissue culture cells. DNA Cell Biol. 1992 Jan-Feb;11(1):83–90. doi: 10.1089/dna.1992.11.83. [DOI] [PubMed] [Google Scholar]
- Shaw W. V. Bacterial resistance to chloramphenicol. Br Med Bull. 1984 Jan;40(1):36–41. doi: 10.1093/oxfordjournals.bmb.a071945. [DOI] [PubMed] [Google Scholar]
- Stokes H. W., Hall R. M. A novel family of potentially mobile DNA elements encoding site-specific gene-integration functions: integrons. Mol Microbiol. 1989 Dec;3(12):1669–1683. doi: 10.1111/j.1365-2958.1989.tb00153.x. [DOI] [PubMed] [Google Scholar]
- Stokes H. W., Hall R. M. Sequence analysis of the inducible chloramphenicol resistance determinant in the Tn1696 integron suggests regulation by translational attenuation. Plasmid. 1991 Jul;26(1):10–19. doi: 10.1016/0147-619x(91)90032-r. [DOI] [PubMed] [Google Scholar]
- Sundström L., Rådström P., Swedberg G., Sköld O. Site-specific recombination promotes linkage between trimethoprim- and sulfonamide resistance genes. Sequence characterization of dhfrV and sulI and a recombination active locus of Tn21. Mol Gen Genet. 1988 Aug;213(2-3):191–201. doi: 10.1007/BF00339581. [DOI] [PubMed] [Google Scholar]
- Surin B. P., Downie J. A. Characterization of the Rhizobium leguminosarum genes nodLMN involved in efficient host-specific nodulation. Mol Microbiol. 1988 Mar;2(2):173–183. doi: 10.1111/j.1365-2958.1988.tb00019.x. [DOI] [PubMed] [Google Scholar]
- Tanaka M., Yamamoto T., Sawai T. Evolution of complex resistance transposons from an ancestral mercury transposon. J Bacteriol. 1983 Mar;153(3):1432–1438. doi: 10.1128/jb.153.3.1432-1438.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennigkeit J., Matzura H. Nucleotide sequence analysis of a chloramphenicol-resistance determinant from Agrobacterium tumefaciens and identification of its gene product. Gene. 1991 Feb 1;98(1):113–116. doi: 10.1016/0378-1119(91)90112-o. [DOI] [PubMed] [Google Scholar]
- Wick M. J., Frank D. W., Storey D. G., Iglewski B. H. Identification of regB, a gene required for optimal exotoxin A yields in Pseudomonas aeruginosa. Mol Microbiol. 1990 Mar;4(3):489–497. doi: 10.1111/j.1365-2958.1990.tb00615.x. [DOI] [PubMed] [Google Scholar]
- Wiedemann B., Meyer J. F., Zühlsdorf M. T. Insertions of resistance genes into Tn21-like transposons. J Antimicrob Chemother. 1986 Oct;18 (Suppl 100):85–92. doi: 10.1093/jac/18.supplement_c.85. [DOI] [PubMed] [Google Scholar]
- Wigley D. B., Derrick J. P., Shaw W. V. The serine acetyltransferase from Escherichia coli. Over-expression, purification and preliminary crystallographic analysis. FEBS Lett. 1990 Dec 17;277(1-2):267–271. doi: 10.1016/0014-5793(90)80862-d. [DOI] [PubMed] [Google Scholar]
- Wohlleben W., Arnold W., Bissonnette L., Pelletier A., Tanguay A., Roy P. H., Gamboa G. C., Barry G. F., Aubert E., Davies J. On the evolution of Tn21-like multiresistance transposons: sequence analysis of the gene (aacC1) for gentamicin acetyltransferase-3-I(AAC(3)-I), another member of the Tn21-based expression cassette. Mol Gen Genet. 1989 Jun;217(2-3):202–208. doi: 10.1007/BF02464882. [DOI] [PubMed] [Google Scholar]



