Abstract
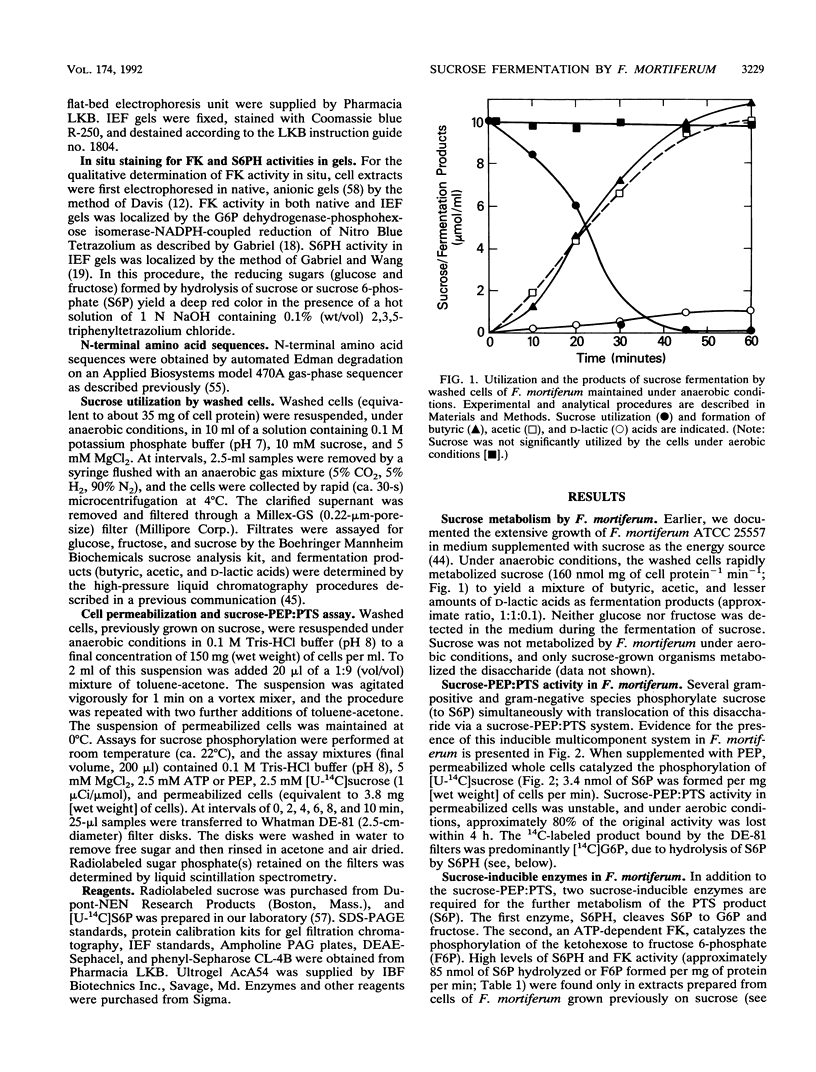
Studies of sucrose utilization by Fusobacterium mortiferum ATCC 25557 have provided the first definitive evidence for phosphoenolpyruvate-dependent sugar:phosphotransferase activity in the family Bacteroidaceae. The phosphoenolpyruvate-dependent sucrose:phosphotransferase system and the two enzymes required for the dissimilation of sucrose 6-phosphate are induced specifically by growth of F. mortiferum on the disaccharide. Monomeric sucrose 6-phosphate hydrolase (M(r), 52,000) and a dimeric ATP-dependent fructokinase (subunit M(r), 32,000) have been purified to electrophoretic homogeneity. The physicochemical and catalytic properties of these enzymes have been examined, and the N-terminal amino acid sequences for both proteins are reported. The characteristics of sucrose 6-phosphate hydrolase and fructokinase from F. mortiferum are compared with the same enzymes from both gram-positive and gram-negative species. Butyric, acetic, and D-lactic acids are the end products of sucrose fermentation by F. mortiferum. A pathway is proposed for the translocation, phosphorylation, and metabolism of sucrose by this anaerobic pathogen.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andrews P. Estimation of the molecular weights of proteins by Sephadex gel-filtration. Biochem J. 1964 May;91(2):222–233. doi: 10.1042/bj0910222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker H. A. Amino acid degradation by anaerobic bacteria. Annu Rev Biochem. 1981;50:23–40. doi: 10.1146/annurev.bi.50.070181.000323. [DOI] [PubMed] [Google Scholar]
- Barker H. A., Kahn J. M., Hedrick L. Pathway of lysine degradation in Fusobacterium nucleatum. J Bacteriol. 1982 Oct;152(1):201–207. doi: 10.1128/jb.152.1.201-207.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blatch G. L., Scholle R. R., Woods D. R. Nucleotide sequence and analysis of the Vibrio alginolyticus sucrose uptake-encoding region. Gene. 1990 Oct 30;95(1):17–23. doi: 10.1016/0378-1119(90)90408-j. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Brook I., Walker R. I. The relationship between Fusobacterium species and other flora in mixed infection. J Med Microbiol. 1986 Mar;21(2):93–100. doi: 10.1099/00222615-21-2-93. [DOI] [PubMed] [Google Scholar]
- Buckel W., Barker H. A. Two pathways of glutamate fermentation by anaerobic bacteria. J Bacteriol. 1974 Mar;117(3):1248–1260. doi: 10.1128/jb.117.3.1248-1260.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chassy B. M., Porter E. V. Initial characterization of sucrose-6-phosphate hydrolase from Streptococcus mutans and its apparent identity with intracellular invertase. Biochem Biophys Res Commun. 1979 Jul 12;89(1):307–314. doi: 10.1016/0006-291x(79)90979-3. [DOI] [PubMed] [Google Scholar]
- Coles R. S., Jr Glucose utilization by resting cells of Fusobacterium polymorphum. Arch Oral Biol. 1977;22(2):87–90. doi: 10.1016/0003-9969(77)90083-8. [DOI] [PubMed] [Google Scholar]
- Cowan P. J., Nagesha H., Leonard L., Howard J. L., Pittard A. J. Characterization of the major promoter for the plasmid-encoded sucrose genes scrY, scrA, and scrB. J Bacteriol. 1991 Dec;173(23):7464–7470. doi: 10.1128/jb.173.23.7464-7470.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DAVIS B. J. DISC ELECTROPHORESIS. II. METHOD AND APPLICATION TO HUMAN SERUM PROTEINS. Ann N Y Acad Sci. 1964 Dec 28;121:404–427. doi: 10.1111/j.1749-6632.1964.tb14213.x. [DOI] [PubMed] [Google Scholar]
- Donkersloot J. A., Thompson J. Simultaneous loss of N5-(carboxyethyl)ornithine synthase, nisin production, and sucrose-fermenting ability by Lactococcus lactis K1. J Bacteriol. 1990 Jul;172(7):4122–4126. doi: 10.1128/jb.172.7.4122-4126.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzink J. L., Socransky S. S. Amino acid utilization by Fusobacterium nucleatum grown in a chemically defined medium. Oral Microbiol Immunol. 1990 Jun;5(3):172–174. doi: 10.1111/j.1399-302x.1990.tb00418.x. [DOI] [PubMed] [Google Scholar]
- Ebner R., Lengeler J. W. DNA sequence of the gene scrA encoding the sucrose transport protein EnzymeII(Scr) of the phosphotransferase system from enteric bacteria: homology of the EnzymeII(Scr) and EnzymeII(Bgl) proteins. Mol Microbiol. 1988 Jan;2(1):9–17. [PubMed] [Google Scholar]
- Fouet A., Klier A., Rapoport G. Nucleotide sequence of the sucrase gene of Bacillus subtilis. Gene. 1986;45(2):221–225. doi: 10.1016/0378-1119(86)90258-1. [DOI] [PubMed] [Google Scholar]
- Franklund C. V., Glass T. L. Glucose uptake by the cellulolytic ruminal anaerobe Bacteroides succinogenes. J Bacteriol. 1987 Feb;169(2):500–506. doi: 10.1128/jb.169.2.500-506.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel O., Wang S. F. Determination of enzymatic activity in polyacrylamide gels. I. Enzymes catalyzing the conversion of nonreducing substrates to reducing products. Anal Biochem. 1969 Mar;27(3):545–554. doi: 10.1016/0003-2697(69)90068-2. [DOI] [PubMed] [Google Scholar]
- George W. L., Kirby B. D., Sutter V. L., Citron D. M., Finegold S. M. Gram-negative anaerobic bacilli: Their role in infection and patterns of susceptibility to antimicrobial agents. II. Little-known Fusobacterium species and miscellaneous genera. Rev Infect Dis. 1981 May-Jun;3(3):599–626. doi: 10.1093/clinids/3.3.599. [DOI] [PubMed] [Google Scholar]
- Gonzalez C. F., Kunka B. S. Transfer of Sucrose-Fermenting Ability and Nisin Production Phenotype among Lactic Streptococci. Appl Environ Microbiol. 1985 Mar;49(3):627–633. doi: 10.1128/aem.49.3.627-633.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardesty C., Ferran C., DiRienzo J. M. Plasmid-mediated sucrose metabolism in Escherichia coli: characterization of scrY, the structural gene for a phosphoenolpyruvate-dependent sucrose phosphotransferase system outer membrane porin. J Bacteriol. 1991 Jan;173(2):449–456. doi: 10.1128/jb.173.2.449-456.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayakawa M., Aoki H., Kuramitsu H. K. Isolation and characterization of the sucrose 6-phosphate hydrolase gene from Streptococcus mutans. Infect Immun. 1986 Sep;53(3):582–586. doi: 10.1128/iai.53.3.582-586.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofstad T. Pathogenicity of anaerobic gram-negative rods: possible mechanisms. Rev Infect Dis. 1984 Mar-Apr;6(2):189–199. doi: 10.1093/clinids/6.2.189. [DOI] [PubMed] [Google Scholar]
- Hylemon P. B., Young J. L., Roadcap R. F., Phibbs P. V., Jr Uptake and incorporation of glucose and mannose by whole cells of Bacteroides thetaiotaomicron. Appl Environ Microbiol. 1977 Nov;34(5):488–494. doi: 10.1128/aem.34.5.488-494.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JACKINS H. C., BARKER H. A. Fermentative processes of the fusiform bacteria. J Bacteriol. 1951 Feb;61(2):101–114. doi: 10.1128/jb.61.2.101-114.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelker N. E., Hanson T. E., Anderson R. L. Alternate pathways of D-fructose metabolism in Aerobacter aerogenes. A specific D-fructokinase and its preferential role in the metabolism of sucrose. J Biol Chem. 1970 Apr 25;245(8):2060–2065. [PubMed] [Google Scholar]
- Kunst F., Pascal M., Lefesant J. A., Walle J., Dedonder R. Purification and some properties of an endocellular sucrase from a constitutive mutant of Bacillus subtilis Marburg 168. Eur J Biochem. 1974 Mar 1;42(2):611–620. doi: 10.1111/j.1432-1033.1974.tb03376.x. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Loesche W. J., Gibbons R. J. Amino acid fermentation by Fusobacterium nucleatum. Arch Oral Biol. 1968 Feb;13(2):191–202. doi: 10.1016/0003-9969(68)90051-4. [DOI] [PubMed] [Google Scholar]
- Lunsford R. D., Macrina F. L. Molecular cloning and characterization of scrB, the structural gene for the Streptococcus mutans phosphoenolpyruvate-dependent sucrose phosphotransferase system sucrose-6-phosphate hydrolase. J Bacteriol. 1986 May;166(2):426–434. doi: 10.1128/jb.166.2.426-434.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maas L. K., Glass T. L. Cellobiose uptake by the cellulolytic ruminal anaerobe Fibrobacter (Bacteroides) succinogenes. Can J Microbiol. 1991 Feb;37(2):141–147. doi: 10.1139/m91-021. [DOI] [PubMed] [Google Scholar]
- Martin S. A., Russell J. B. Phosphoenolpyruvate-dependent phosphorylation of hexoses by ruminal bacteria: evidence for the phosphotransferase transport system. Appl Environ Microbiol. 1986 Dec;52(6):1348–1352. doi: 10.1128/aem.52.6.1348-1352.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter E. V., Chassy B. M., Holmlund C. E. Partial purification and properties of a mannofructokinase from Streptococcus mutans SL-1. Infect Immun. 1980 Oct;30(1):43–50. doi: 10.1128/iai.30.1.43-50.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postma P. W., Lengeler J. W. Phosphoenolpyruvate:carbohydrate phosphotransferase system of bacteria. Microbiol Rev. 1985 Sep;49(3):232–269. doi: 10.1128/mr.49.3.232-269.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robrish S. A., Oliver C., Thompson J. Amino acid-dependent transport of sugars by Fusobacterium nucleatum ATCC 10953. J Bacteriol. 1987 Sep;169(9):3891–3897. doi: 10.1128/jb.169.9.3891-3897.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robrish S. A., Oliver C., Thompson J. Sugar metabolism by fusobacteria: regulation of transport, phosphorylation, and polymer formation by Fusobacterium mortiferum ATCC 25557. Infect Immun. 1991 Dec;59(12):4547–4554. doi: 10.1128/iai.59.12.4547-4554.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robrish S. A., Thompson J. Regulation of fructose metabolism and polymer synthesis by Fusobacterium nucleatum ATCC 10953. J Bacteriol. 1990 Oct;172(10):5714–5723. doi: 10.1128/jb.172.10.5714-5723.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y., Kuramitsu H. K. Sequence analysis of the Streptococcus mutans scrB gene. Infect Immun. 1988 Aug;56(8):1956–1960. doi: 10.1128/iai.56.8.1956-1960.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid K., Ebner R., Altenbuchner J., Schmitt R., Lengeler J. W. Plasmid-mediated sucrose metabolism in Escherichia coli K12: mapping of the scr genes of pUR400. Mol Microbiol. 1988 Jan;2(1):1–8. doi: 10.1111/j.1365-2958.1988.tb00001.x. [DOI] [PubMed] [Google Scholar]
- Schmid K., Schupfner M., Schmitt R. Plasmid-mediated uptake and metabolism of sucrose by Escherichia coli K-12. J Bacteriol. 1982 Jul;151(1):68–76. doi: 10.1128/jb.151.1.68-76.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholle R. R., Robb S. M., Robb F. T., Woods D. R. Nucleotide sequence and analysis of the Vibrio alginolyticus sucrase gene (scrB). Gene. 1989 Aug 1;80(1):49–56. doi: 10.1016/0378-1119(89)90249-7. [DOI] [PubMed] [Google Scholar]
- Scholle R. R., Steffen H. E., Goodman H. J., Woods D. R. Expression and regulation of a Bacteroides fragilis sucrose utilization system cloned in Escherichia coli. Appl Environ Microbiol. 1990 Jun;56(6):1944–1948. doi: 10.1128/aem.56.6.1944-1948.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastian J., Asensio C. Purification and properties of the mannokinase from Escherichia coli. Arch Biochem Biophys. 1972 Jul;151(1):227–233. doi: 10.1016/0003-9861(72)90492-4. [DOI] [PubMed] [Google Scholar]
- Sprenger G. A., Lengeler J. W. Analysis of sucrose catabolism in Klebsiella pneumoniae and in Scr+ derivatives of Escherichia coli K12. J Gen Microbiol. 1988 Jun;134(6):1635–1644. doi: 10.1099/00221287-134-6-1635. [DOI] [PubMed] [Google Scholar]
- Steele J. L., McKay L. L. Partial characterization of the genetic basis for sucrose metabolism and nisin production in Streptococcus lactis. Appl Environ Microbiol. 1986 Jan;51(1):57–64. doi: 10.1128/aem.51.1.57-64.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J., Chassy B. M. Uptake and metabolism of sucrose by Streptococcus lactis. J Bacteriol. 1981 Aug;147(2):543–551. doi: 10.1128/jb.147.2.543-551.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J. N5-(L-1-carboxyethyl)-L-ornithine:NADP+ oxidoreductase from Streptococcus lactis. Purification and partial characterization. J Biol Chem. 1989 Jun 5;264(16):9592–9601. [PubMed] [Google Scholar]
- Thompson J., Nguyen N. Y., Sackett D. L., Donkersloot J. A. Transposon-encoded sucrose metabolism in Lactococcus lactis. Purification of sucrose-6-phosphate hydrolase and genetic linkage to N5-(L-1-carboxyethyl)-L-ornithine synthase in strain K1. J Biol Chem. 1991 Aug 5;266(22):14573–14579. [PubMed] [Google Scholar]
- Thompson J., Sackett D. L., Donkersloot J. A. Purification and properties of fructokinase I from Lactococcus lactis. Localization of scrK on the sucrose-nisin transposon Tn5306. J Biol Chem. 1991 Nov 25;266(33):22626–22633. [PubMed] [Google Scholar]
- Wehmeier U., Sprenger G. A., Lengeler J. W. The use of lambda plac-Mu hybrid phages in Klebsiella pneumoniae and the isolation of stable Hfr strains. Mol Gen Genet. 1989 Feb;215(3):529–536. doi: 10.1007/BF00427052. [DOI] [PubMed] [Google Scholar]



