Abstract
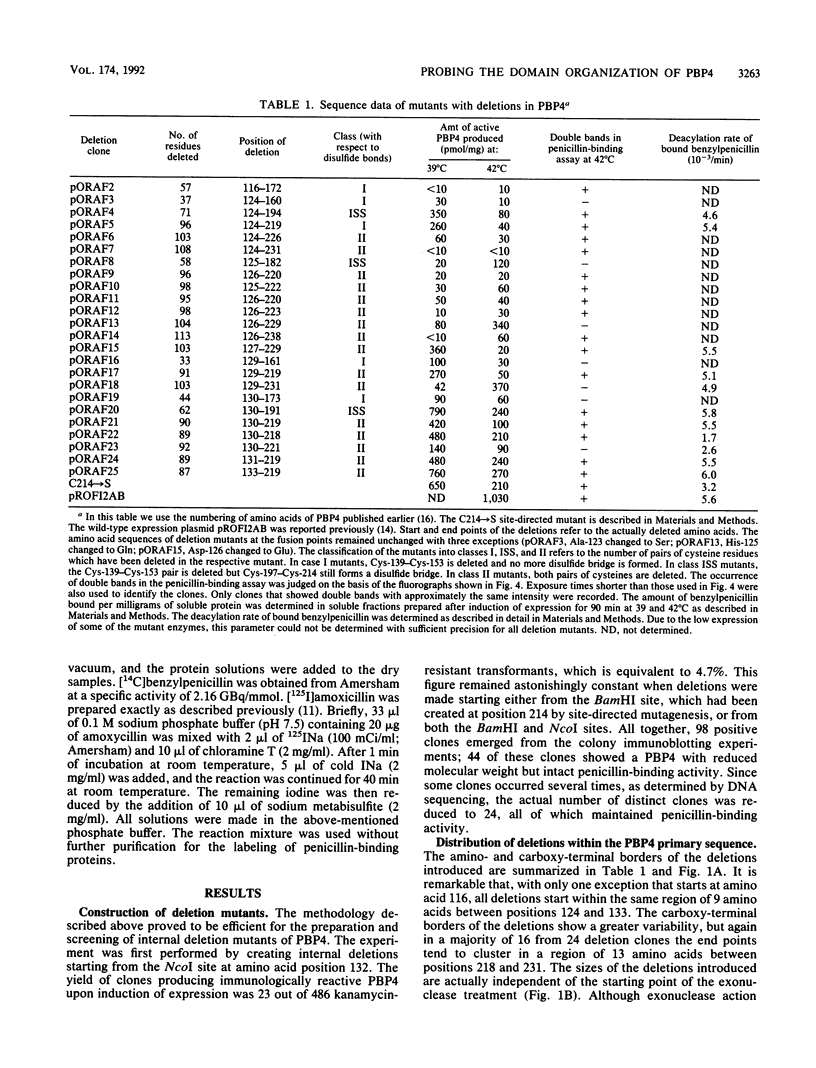
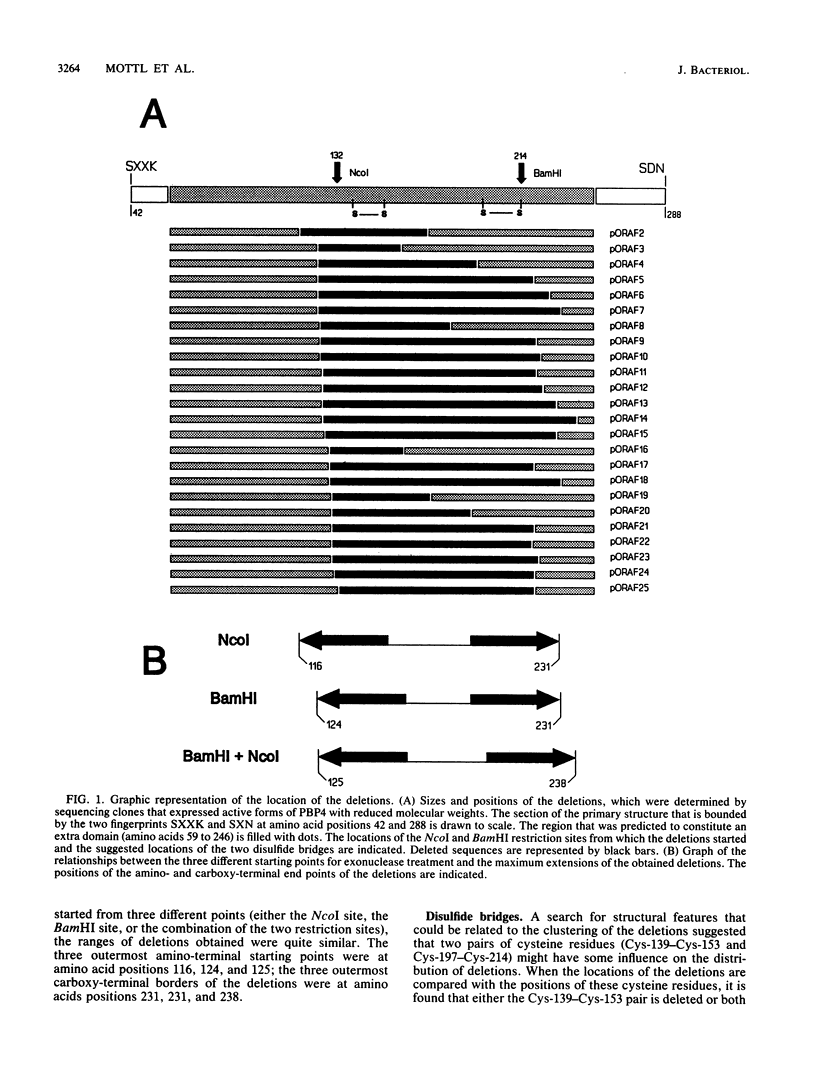
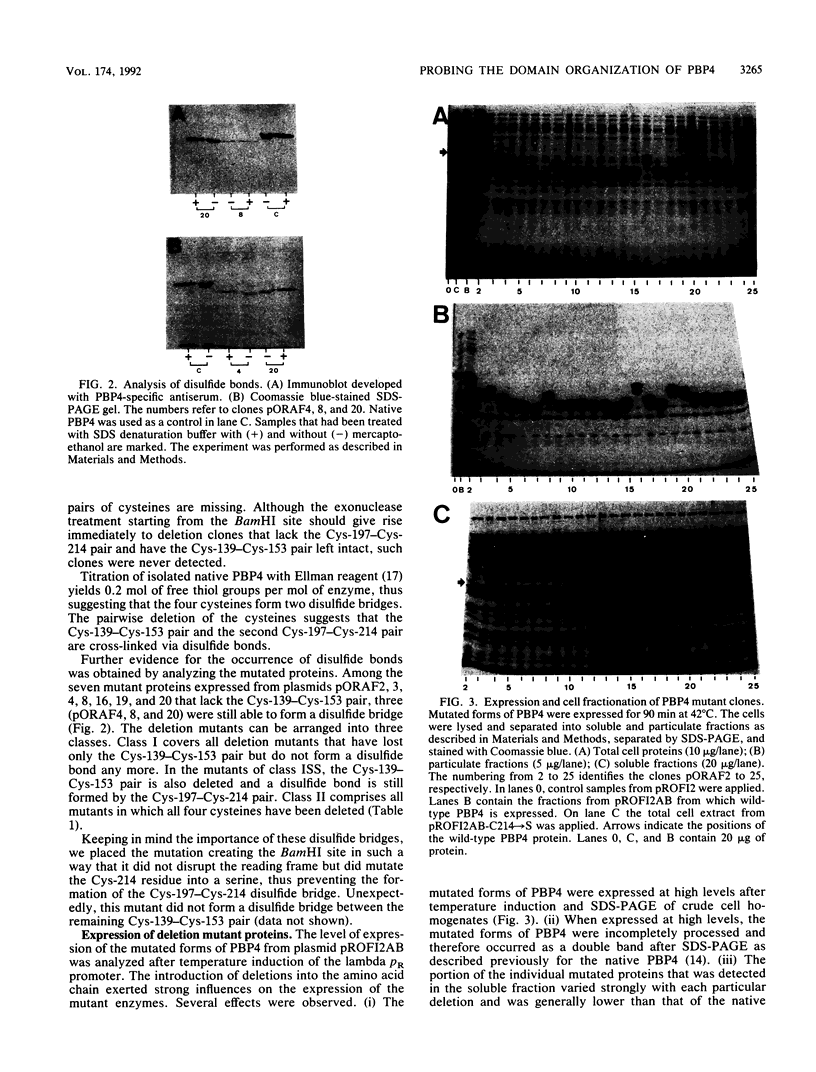
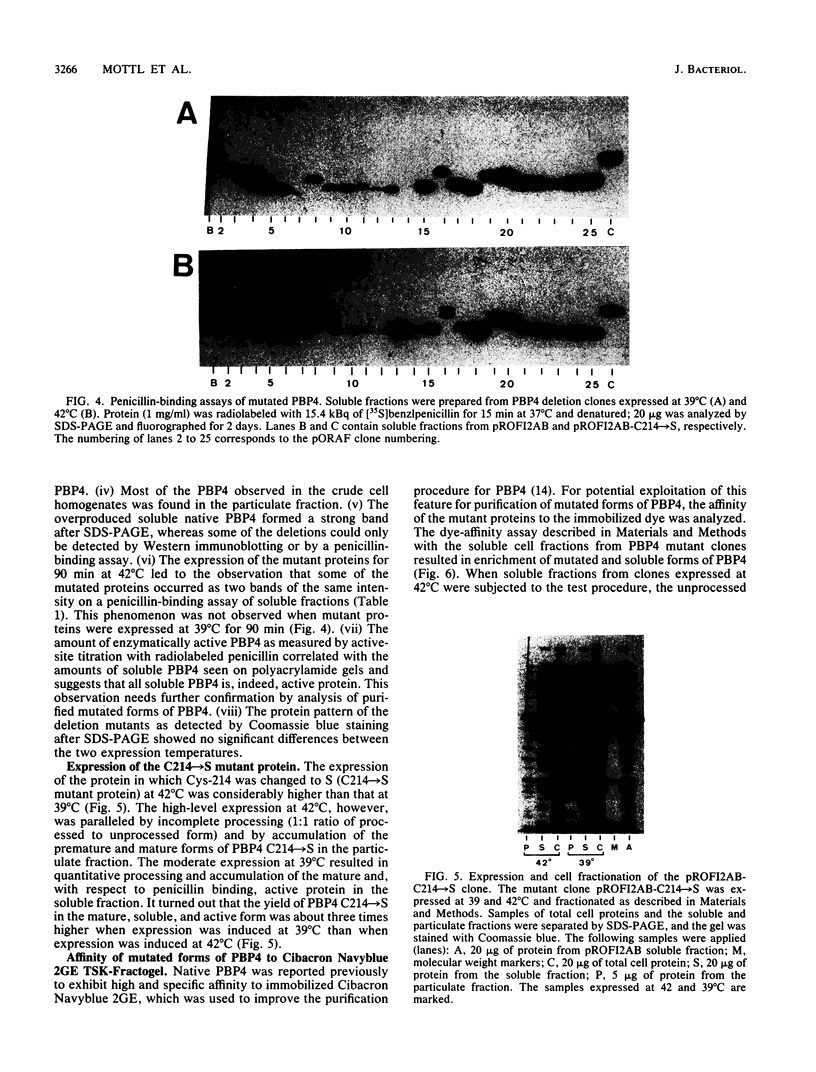
It was suggested previously that the primary structure of penicillin-binding protein 4 (PBP4) is new and unique among proteins that interact with penicillin. Our proposal that PBP4 carries an additional domain, located between the active-site fingerprints SXXK and SXN, was investigated by mutational deletion analysis. A clustered set of internal deletions was created in this region by exonuclease treatment of the dacB coding DNA, starting from two internal restriction sites. PBP4 mutants carrying internal deletions were selected by screening for immunoreactive forms of PBP4 with reduced molecular weight that were still active with respect to penicillin binding. DNA sequencing revealed 24 distinct PBP4 mutants with internal deletions ranging from 37 to 113 amino acids. The amino- and carboxy-terminal end points of the deletions were not randomly distributed but tended to cluster in certain areas. Overproduction of the individual mutated forms of PBP4 resulted in accumulation of the major portion of the proteins in the particulate cell fraction. The yield of soluble and active mutated forms of the protein was reduced from below 1% to 79% of the level obtained for the native protein. The deletions that were introduced had minor effects on the deacylation rate of bound benzylpenicillin. Two pairs of cysteine residues (Cys-139-Cys-153 and Cys-197-Cys-214) that are located in the deletable region may form disulfide bridges.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allore R. J., Barber B. H. A recommendation for visualizing disulfide bonding by one-dimensional sodium dodecyl sulfate--polyacrylamide gel electrophoresis. Anal Biochem. 1984 Mar;137(2):523–527. doi: 10.1016/0003-2697(84)90121-0. [DOI] [PubMed] [Google Scholar]
- Chamberlain J. P. Fluorographic detection of radioactivity in polyacrylamide gels with the water-soluble fluor, sodium salicylate. Anal Biochem. 1979 Sep 15;98(1):132–135. doi: 10.1016/0003-2697(79)90716-4. [DOI] [PubMed] [Google Scholar]
- Hakenbeck R., Messer W. Activity of murein hydrolases in synchronized cultures of Escherichia coli. J Bacteriol. 1977 Mar;129(3):1239–1244. doi: 10.1128/jb.129.3.1239-1244.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzberg O., Moult J. Bacterial resistance to beta-lactam antibiotics: crystal structure of beta-lactamase from Staphylococcus aureus PC1 at 2.5 A resolution. Science. 1987 May 8;236(4802):694–701. doi: 10.1126/science.3107125. [DOI] [PubMed] [Google Scholar]
- Korat B., Mottl H., Keck W. Penicillin-binding protein 4 of Escherichia coli: molecular cloning of the dacB gene, controlled overexpression, and alterations in murein composition. Mol Microbiol. 1991 Mar;5(3):675–684. doi: 10.1111/j.1365-2958.1991.tb00739.x. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Masson J. M., Labia R. Synthesis of a 125I-radiolabeled penicillin for penicillin-binding proteins studies. Anal Biochem. 1983 Jan;128(1):164–168. doi: 10.1016/0003-2697(83)90357-3. [DOI] [PubMed] [Google Scholar]
- Meissner P. S., Sisk W. P., Berman M. L. Bacteriophage lambda cloning system for the construction of directional cDNA libraries. Proc Natl Acad Sci U S A. 1987 Jun;84(12):4171–4175. doi: 10.1073/pnas.84.12.4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Mottl H., Keck W. Purification of penicillin-binding protein 4 of Escherichia coli as a soluble protein by dye-affinity chromatography. Eur J Biochem. 1991 Sep 15;200(3):767–773. doi: 10.1111/j.1432-1033.1991.tb16243.x. [DOI] [PubMed] [Google Scholar]
- Mottl H., Terpstra P., Keck W. Penicillin-binding protein 4 of Escherichia coli shows a novel type of primary structure among penicillin-interacting proteins. FEMS Microbiol Lett. 1991 Mar 1;62(2-3):213–220. doi: 10.1016/0378-1097(91)90160-c. [DOI] [PubMed] [Google Scholar]
- Riddles P. W., Blakeley R. L., Zerner B. Reassessment of Ellman's reagent. Methods Enzymol. 1983;91:49–60. doi: 10.1016/s0076-6879(83)91010-8. [DOI] [PubMed] [Google Scholar]
- Spratt B. G., Cromie K. D. Penicillin-binding proteins of gram-negative bacteria. Rev Infect Dis. 1988 Jul-Aug;10(4):699–711. doi: 10.1093/clinids/10.4.699. [DOI] [PubMed] [Google Scholar]
- Stanley K. K. Solubilization and immune-detection of beta-galactosidase hybrid proteins carrying foreign antigenic determinants. Nucleic Acids Res. 1983 Jun 25;11(12):4077–4092. doi: 10.1093/nar/11.12.4077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuomanen E., Lindquist S., Sande S., Galleni M., Light K., Gage D., Normark S. Coordinate regulation of beta-lactamase induction and peptidoglycan composition by the amp operon. Science. 1991 Jan 11;251(4990):201–204. doi: 10.1126/science.1987637. [DOI] [PubMed] [Google Scholar]
- Witholt B., Boekhout M., Brock M., Kingma J., Heerikhuizen H. V., Leij L. D. An efficient and reproducible procedure for the formation of spheroplasts from variously grown Escherichia coli. Anal Biochem. 1976 Jul;74(1):160–170. doi: 10.1016/0003-2697(76)90320-1. [DOI] [PubMed] [Google Scholar]







