Abstract
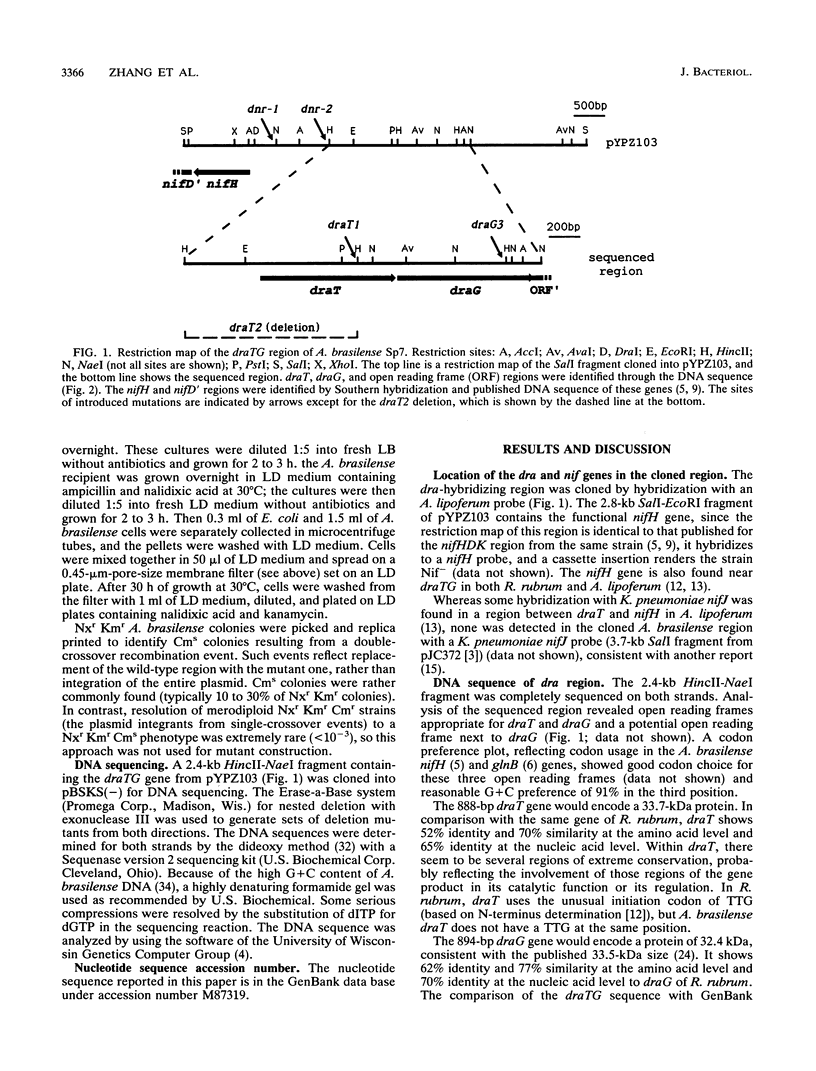
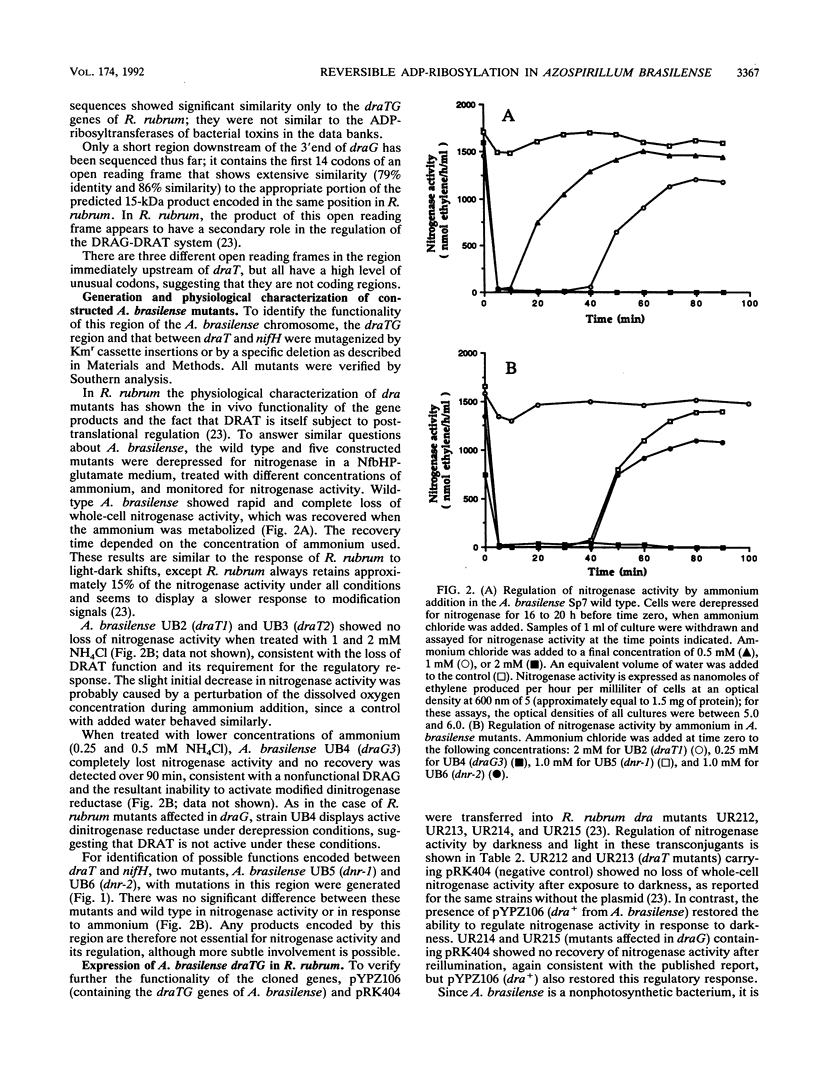
The Azospirillum brasilense draT gene, encoding dinitrogenase reductase ATP-ribosyltransferase, and draG gene, encoding dinitrogenase reductase activating glycohydrolase, were cloned and sequenced. Two genes were contiguous on the A. brasilense chromosome and showed extensive similarity to the same genes from Rhodospirillum rubrum. Analysis of mutations introduced into the dra region on the A. brasilense chromosome showed that mutants affected in draT were incapable of regulating nitrogenase activity in response to ammonium. In contrast, a mutant with an insertion in draG was still capable of ADP-ribosylating dinitrogenase reductase in response to ammonium but was no longer able to recover activity after ammonium depletion. Plasmid-borne draTG genes from A. brasilense were introduced into dra mutants of R. rubrum and restored these mutants to an apparently wild-type phenotype. It is particularly interesting that dra mutants of R. rubrum containing draTG of A. brasilense can respond to darkness and light, since A. brasilense is a nonphotosynthetic bacterium and its dra system does not normally possess that regulatory response. The nifH gene of A. brasilense, encoding dinitrogenase reductase (the substrate of dinitrogenase reductase ADP-ribosyltransferase and dinitrogenase reductase-activating glycohydrolase), is located 1.9 kb from the start of draT and is divergently transcribed. Two insertion mutations in the region between draT and nifH showed no significant effect on nitrogenase activity or its regulation.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Burris R. H. Nitrogenases. J Biol Chem. 1991 May 25;266(15):9339–9342. [PubMed] [Google Scholar]
- Collins J. J., Roberts G. P., Brill W. J. Posttranscriptional control of Klebsiella pneumoniae nif mRNA stability by the nifL product. J Bacteriol. 1986 Oct;168(1):173–178. doi: 10.1128/jb.168.1.173-178.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditta G., Schmidhauser T., Yakobson E., Lu P., Liang X. W., Finlay D. R., Guiney D., Helinski D. R. Plasmids related to the broad host range vector, pRK290, useful for gene cloning and for monitoring gene expression. Plasmid. 1985 Mar;13(2):149–153. doi: 10.1016/0147-619x(85)90068-x. [DOI] [PubMed] [Google Scholar]
- Fani R., Allotta G., Bazzicalupo M., Ricci F., Schipani C., Polsinelli M. Nucleotide sequence of the gene encoding the nitrogenase iron protein (nifH) of Azospirillum brasilense and identification of a region controlling nifH transcription. Mol Gen Genet. 1989 Dec;220(1):81–87. doi: 10.1007/BF00260860. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. "A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity". Addendum. Anal Biochem. 1984 Feb;137(1):266–267. doi: 10.1016/0003-2697(84)90381-6. [DOI] [PubMed] [Google Scholar]
- Figurski D. H., Helinski D. R. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzmaurice W. P., Saari L. L., Lowery R. G., Ludden P. W., Roberts G. P. Genes coding for the reversible ADP-ribosylation system of dinitrogenase reductase from Rhodospirillum rubrum. Mol Gen Genet. 1989 Aug;218(2):340–347. doi: 10.1007/BF00331287. [DOI] [PubMed] [Google Scholar]
- Fu H. A., Fitzmaurice W. P., Roberts G. P., Burris R. H. Cloning and expression of draTG genes from Azospirillum lipoferum. Gene. 1990 Jan 31;86(1):95–98. doi: 10.1016/0378-1119(90)90118-b. [DOI] [PubMed] [Google Scholar]
- Fu H. A., Hartmann A., Lowery R. G., Fitzmaurice W. P., Roberts G. P., Burris R. H. Posttranslational regulatory system for nitrogenase activity in Azospirillum spp. J Bacteriol. 1989 Sep;171(9):4679–4685. doi: 10.1128/jb.171.9.4679-4685.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galimand M., Perroud B., Delorme F., Paquelin A., Vieille C., Bozouklian H., Elmerich C. Identification of DNA regions homologous to nitrogen fixation genes nifE, nifUS and fixABC in Azospirillum brasilense Sp7. J Gen Microbiol. 1989 May;135(5):1047–1059. doi: 10.1099/00221287-135-5-1047. [DOI] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983 Jun 5;166(4):557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Hartmann A., Fu H., Burris R. H. Regulation of nitrogenase activity by ammonium chloride in Azospirillum spp. J Bacteriol. 1986 Mar;165(3):864–870. doi: 10.1128/jb.165.3.864-870.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanemoto R. H., Ludden P. W. Amino acid concentrations in Rhodospirillum rubrum during expression and switch-off of nitrogenase activity. J Bacteriol. 1987 Jul;169(7):3035–3043. doi: 10.1128/jb.169.7.3035-3043.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanemoto R. H., Ludden P. W. Effect of ammonia, darkness, and phenazine methosulfate on whole-cell nitrogenase activity and Fe protein modification in Rhodospirillum rubrum. J Bacteriol. 1984 May;158(2):713–720. doi: 10.1128/jb.158.2.713-720.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laane C., Krone W., Konings W., Haaker H., Veeger C. Short-term effect of ammonium chloride on nitrogen fixation by Azotobacter vinelandii and by bacteroids of Rhizobium leguminosarum. Eur J Biochem. 1980 Jan;103(1):39–46. doi: 10.1111/j.1432-1033.1980.tb04286.x. [DOI] [PubMed] [Google Scholar]
- Lehman L. J., Roberts G. P. Identification of an alternative nitrogenase system in Rhodospirillum rubrum. J Bacteriol. 1991 Sep;173(18):5705–5711. doi: 10.1128/jb.173.18.5705-5711.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. D., Hu C. Z., Yoch D. C. Changes in amino acid and nucleotide pools of Rhodospirillum rubrum during switch-off of nitrogenase activity initiated by NH4+ or darkness. J Bacteriol. 1987 Jan;169(1):231–237. doi: 10.1128/jb.169.1.231-237.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J. H., Nielsen G. M., Lies D. P., Burris R. H., Roberts G. P., Ludden P. W. Mutations in the draT and draG genes of Rhodospirillum rubrum result in loss of regulation of nitrogenase by reversible ADP-ribosylation. J Bacteriol. 1991 Nov;173(21):6903–6909. doi: 10.1128/jb.173.21.6903-6909.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludden P. W., Roberts G. P. Regulation of nitrogenase activity by reversible ADP ribosylation. Curr Top Cell Regul. 1989;30:23–56. doi: 10.1016/b978-0-12-152830-0.50004-9. [DOI] [PubMed] [Google Scholar]
- Paul T. D., Ludden P. W. Adenine nucleotide levels in Rhodospirillum rubrum during switch-off of whole-cell nitrogenase activity. Biochem J. 1984 Dec 15;224(3):961–969. doi: 10.1042/bj2240961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed K. C., Mann D. A. Rapid transfer of DNA from agarose gels to nylon membranes. Nucleic Acids Res. 1985 Oct 25;13(20):7207–7221. doi: 10.1093/nar/13.20.7207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarrand J. J., Krieg N. R., Döbereiner J. A taxonomic study of the Spirillum lipoferum group, with descriptions of a new genus, Azospirillum gen. nov. and two species, Azospirillum lipoferum (Beijerinck) comb. nov. and Azospirillum brasilense sp. nov. Can J Microbiol. 1978 Aug;24(8):967–980. doi: 10.1139/m78-160. [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Zumft W. G., Castillo F. Regulatory properties of the nitrogenase from Rhodopseudomonas palustris. Arch Microbiol. 1978 Apr 27;117(1):53–60. doi: 10.1007/BF00689351. [DOI] [PubMed] [Google Scholar]
- de Zamaroczy M., Delorme F., Elmerich C. Characterization of three different nitrogen-regulated promoter regions for the expression of glnB and glnA in Azospirillum brasilense. Mol Gen Genet. 1990 Dec;224(3):421–430. doi: 10.1007/BF00262437. [DOI] [PubMed] [Google Scholar]
- de Zamaroczy M., Delorme F., Elmerich C. Regulation of transcription and promoter mapping of the structural genes for nitrogenase (nifHDK) of Azospirillum brasilense Sp7. Mol Gen Genet. 1989 Dec;220(1):88–94. doi: 10.1007/BF00260861. [DOI] [PubMed] [Google Scholar]


