Abstract
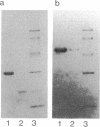
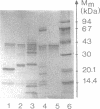
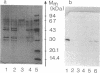
Five extracellular chitinases of 20.5, 30, 47, 70, and 92 kDa purified from the culture filtrate of Streptomyces olivaceoviridis ATCC 11238 differed in their sequences at the amino termini of the protein chains. In the native state, the chitinases were found to be resistant to proteolysis by trypsin, papain, and Staphylococcus aureus V8 protease. The latter produced several fragments of identical molecular mass from chitinases denaturated with sodium dodecyl sulfate. Five proteases were detected in the protein concentrate from the culture filtrate, and two of them showing ability to cleave chitinases in the native state were purified. One, a protease of 42 kDa, released a 30-kDa protein from the 70-kDa chitinase that reacts with anti-30 kDa chitinase antibodies; the other, a protease of 29 kDa, split the 30-kDa chitinase into 20.5-, 18-, and 16-kDa fragments. From these results, it was deduced that the 70-kDa chitinase is the precursor protein of the 30- and 20.5-kDa chitinases.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Cleveland D. W., Fischer S. G., Kirschner M. W., Laemmli U. K. Peptide mapping by limited proteolysis in sodium dodecyl sulfate and analysis by gel electrophoresis. J Biol Chem. 1977 Feb 10;252(3):1102–1106. [PubMed] [Google Scholar]
- Dygert S., Li L. H., Florida D., Thoma J. A. Determination of reducing sugar with improved precision. Anal Biochem. 1965 Dec;13(3):367–374. doi: 10.1016/0003-2697(65)90327-1. [DOI] [PubMed] [Google Scholar]
- Garcia-Gonzalez M. D., Martin J. F., Vigal T., Liras P. Characterization, expression in Streptomyces lividans, and processing of the amylase of Streptomyces griseus IMRU 3570: two different amylases are derived from the same gene by an intracellular processing mechanism. J Bacteriol. 1991 Apr;173(8):2451–2458. doi: 10.1128/jb.173.8.2451-2458.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara S., Yamamura Y., Fujii Y., Mega T., Ikenaka T. Purification and characterization of chitinase produced by Streptomyces erythraeus. J Biochem. 1989 Mar;105(3):484–489. doi: 10.1093/oxfordjournals.jbchem.a122691. [DOI] [PubMed] [Google Scholar]
- Hewick R. M., Hunkapiller M. W., Hood L. E., Dreyer W. J. A gas-liquid solid phase peptide and protein sequenator. J Biol Chem. 1981 Aug 10;256(15):7990–7997. [PubMed] [Google Scholar]
- Hopwood D. A. Genetic analysis and genome structure in Streptomyces coelicolor. Bacteriol Rev. 1967 Dec;31(4):373–403. doi: 10.1128/br.31.4.373-403.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lüscher B., Brizuela L., Beach D., Eisenman R. N. A role for the p34cdc2 kinase and phosphatases in the regulation of phosphorylation and disassembly of lamin B2 during the cell cycle. EMBO J. 1991 Apr;10(4):865–875. doi: 10.1002/j.1460-2075.1991.tb08019.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsutomi M., Ohtakara A., Fukamizo T., Goto S. Action pattern of Aeromonas hydrophila chitinase on partially N-acetylated chitosan. Agric Biol Chem. 1990 Apr;54(4):871–877. [PubMed] [Google Scholar]
- Nakada T., Kubota M., Sakai S., Tsujisaka Y. Purification and characterization of two forms of maltotetraose-forming amylase from Pseudomonas stutzeri. Agric Biol Chem. 1990 Mar;54(3):737–743. [PubMed] [Google Scholar]
- Pel R., van den Wijngaard A. J., Epping E., Gottschal J. C. Comparison of the chitinolytic properties of Clostridium sp. strain 9.1 and a chitin-degrading bacterium from the intestinal tract of the plaice, Pleuronectes platessa (L.). J Gen Microbiol. 1990 Apr;136(4):695–704. doi: 10.1099/00221287-136-4-695. [DOI] [PubMed] [Google Scholar]
- Robbins P. W., Albright C., Benfield B. Cloning and expression of a Streptomyces plicatus chitinase (chitinase-63) in Escherichia coli. J Biol Chem. 1988 Jan 5;263(1):443–447. [PubMed] [Google Scholar]
- Takahashi T., Tsuchida Y., Irie M. Isolation of two inactive fragments of a Rhizopus sp. glucoamylase: relationship among three forms of the enzyme and the isolated fragments. J Biochem. 1982 Nov;92(5):1623–1633. doi: 10.1093/oxfordjournals.jbchem.a134088. [DOI] [PubMed] [Google Scholar]
- Takayanagi T., Ajisaka K., Takiguchi Y., Shimahara K. Isolation and characterization of thermostable chitinases from Bacillus licheniformis X-7u. Biochim Biophys Acta. 1991 Jul 12;1078(3):404–410. doi: 10.1016/0167-4838(91)90163-t. [DOI] [PubMed] [Google Scholar]
- Tarentino A. L., Maley F. Purification and properties of an endo-beta-N-acetylglucosaminidase from Streptomyces griseus. J Biol Chem. 1974 Feb 10;249(3):811–817. [PubMed] [Google Scholar]
- Uozumi N., Sakurai K., Sasaki T., Takekawa S., Yamagata H., Tsukagoshi N., Udaka S. A single gene directs synthesis of a precursor protein with beta- and alpha-amylase activities in Bacillus polymyxa. J Bacteriol. 1989 Jan;171(1):375–382. doi: 10.1128/jb.171.1.375-382.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T., Oyanagi W., Suzuki K., Tanaka H. Chitinase system of Bacillus circulans WL-12 and importance of chitinase A1 in chitin degradation. J Bacteriol. 1990 Jul;172(7):4017–4022. doi: 10.1128/jb.172.7.4017-4022.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]