Abstract
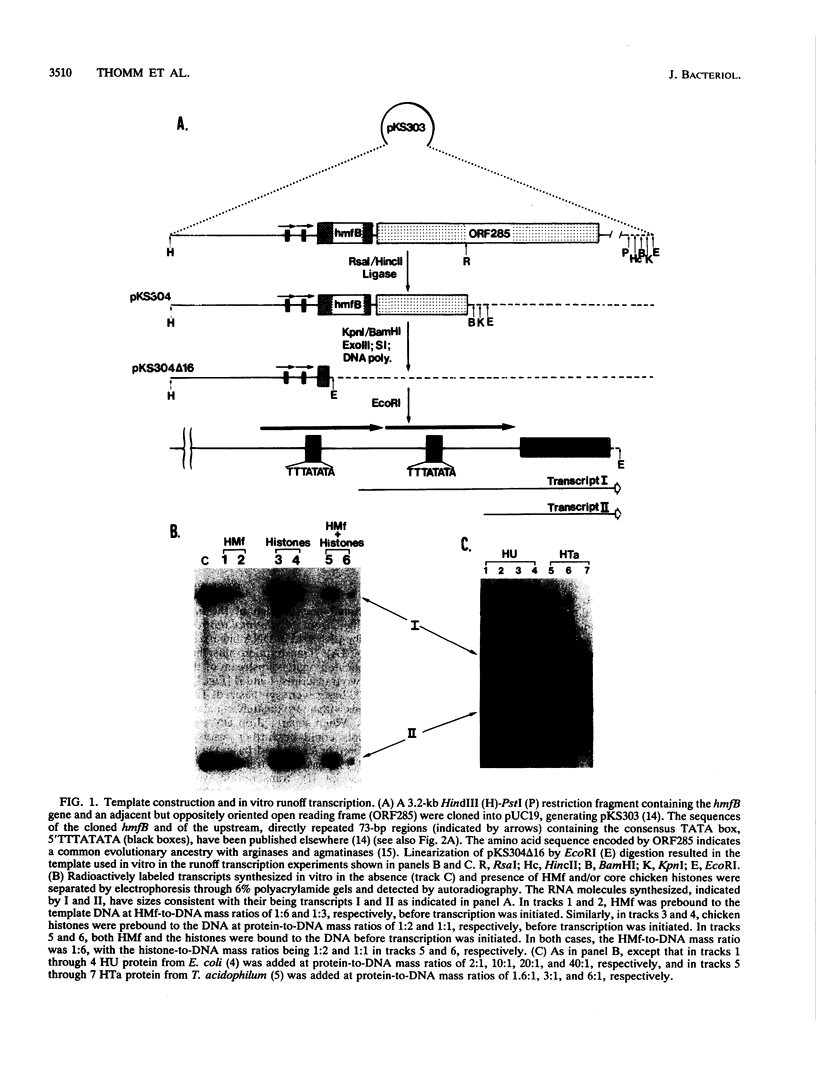
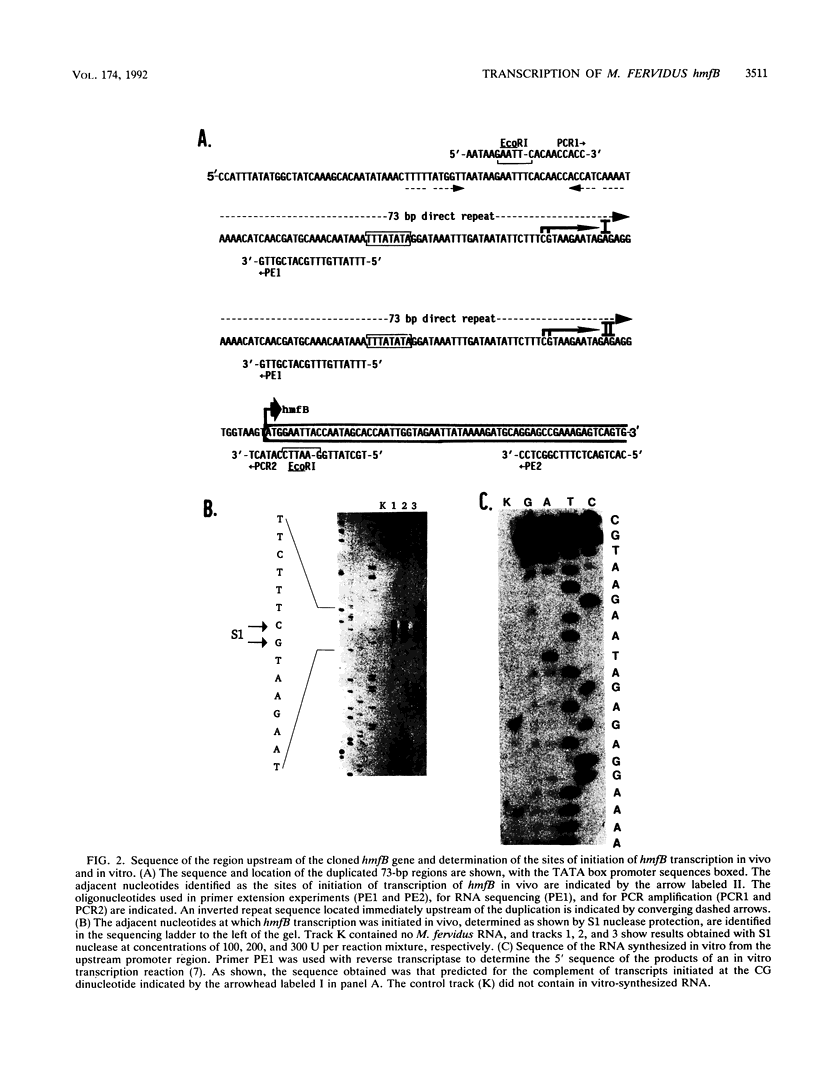
Immediately upstream of the hmfB gene, in a DNA fragment cloned from Methanothermus fervidus, are two identical tandemly repeated copies of a 73-bp sequence that contain the sequence 5'TTTATATA, which conforms precisely to the consensus TATA box element proposed for methanogen promoters. By using this duplicated region as the template DNA and a cell-free transcription system derived from Methanococcus thermolithotrophicus, transcription in vitro was found to initiate at two identical sites 73 bp apart, each 25 bp downstream from a TATA box, thus providing strong evidence for the functional conservation of this transcriptional signal in two phylogenetically very diverse methanogens. Transcription of the hmfB gene in vivo in M. fervidus was found to occur at only one of these sites, and consistent with this observation, recloning and sequencing of this intergenic region after its amplification by the polymerase chain reaction demonstrated that the genome of M. fervidus contains only one copy of the 73-bp sequence upstream of the hmfB gene. Since the second copy of the 73-bp sequence, presumably generated artifactually during the original hmfB cloning, functioned equally well as a promoter in the M. thermolithotrophicus transcription system, all information needed by the heterologous RNA polymerase to initiate transcription accurately in vitro must be present within this sequence. The hmfB gene encodes HMf-2, one of the two subunits of HMf, an abundant DNA binding protein in M. fervidus which binds to DNA molecules in vitro, forming nucleosomelike structures. Cell-free transcription was inhibited by adding HMf or eucaryotic core histones at protein-to-DNA mass ratios of 0.3:1 and 1:1, respectively, whereas the archael histonelike protein HTa from Thermoplasma acidophilum inhibited transcription in vitro only at much higher protein-to-DNA mass ratios and the bacterial histonelike protein HU from Escherichia coli had no detectable effect on transcription.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berk A. J., Sharp P. A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977 Nov;12(3):721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- Brown J. W., Daniels C. J., Reeve J. N. Gene structure, organization, and expression in archaebacteria. Crit Rev Microbiol. 1989;16(4):287–338. doi: 10.3109/10408418909105479. [DOI] [PubMed] [Google Scholar]
- Brown J. W., Thomm M., Beckler G. S., Frey G., Stetter K. O., Reeve J. N. An archaebacterial RNA polymerase binding site and transcription initiation of the hisA gene in Methanococcus vannielii. Nucleic Acids Res. 1988 Jan 11;16(1):135–150. doi: 10.1093/nar/16.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broyles S. S., Pettijohn D. E. Interaction of the Escherichia coli HU protein with DNA. Evidence for formation of nucleosome-like structures with altered DNA helical pitch. J Mol Biol. 1986 Jan 5;187(1):47–60. doi: 10.1016/0022-2836(86)90405-5. [DOI] [PubMed] [Google Scholar]
- DeLange R. J., Green G. R., Searcy D. G. A histone-like protein (HTa) from Thermoplasma acidophilum. I. Purification and properties. J Biol Chem. 1981 Jan 25;256(2):900–904. [PubMed] [Google Scholar]
- Elgin S. C. The formation and function of DNase I hypersensitive sites in the process of gene activation. J Biol Chem. 1988 Dec 25;263(36):19259–19262. [PubMed] [Google Scholar]
- Frey G., Thomm M., Brüdigam B., Gohl H. P., Hausner W. An archaebacterial cell-free transcription system. The expression of tRNA genes from Methanococcus vannielii is mediated by a transcription factor. Nucleic Acids Res. 1990 Mar 25;18(6):1361–1367. doi: 10.1093/nar/18.6.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas E. S., Brown J. W., Daniels C. J., Reeve J. N. Genes encoding the 7S RNA and tRNA(Ser) are linked to one of the two rRNA operons in the genome of the extremely thermophilic archaebacterium Methanothermus fervidus. Gene. 1990 May 31;90(1):51–59. doi: 10.1016/0378-1119(90)90438-w. [DOI] [PubMed] [Google Scholar]
- Hausner W., Frey G., Thomm M. Control regions of an archaeal gene. A TATA box and an initiator element promote cell-free transcription of the tRNA(Val) gene of Methanococcus vannielii. J Mol Biol. 1991 Dec 5;222(3):495–508. doi: 10.1016/0022-2836(91)90492-o. [DOI] [PubMed] [Google Scholar]
- Jones W. J., Nagle D. P., Jr, Whitman W. B. Methanogens and the diversity of archaebacteria. Microbiol Rev. 1987 Mar;51(1):135–177. doi: 10.1128/mr.51.1.135-177.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musgrave D. R., Sandman K. M., Reeve J. N. DNA binding by the archaeal histone HMf results in positive supercoiling. Proc Natl Acad Sci U S A. 1991 Dec 1;88(23):10397–10401. doi: 10.1073/pnas.88.23.10397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter W. D., Palm P., Zillig W. Analysis of transcription in the archaebacterium Sulfolobus indicates that archaebacterial promoters are homologous to eukaryotic pol II promoters. Nucleic Acids Res. 1988 Jan 11;16(1):1–19. doi: 10.1093/nar/16.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandman K., Krzycki J. A., Dobrinski B., Lurz R., Reeve J. N. HMf, a DNA-binding protein isolated from the hyperthermophilic archaeon Methanothermus fervidus, is most closely related to histones. Proc Natl Acad Sci U S A. 1990 Aug;87(15):5788–5791. doi: 10.1073/pnas.87.15.5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M. B. More than just "histone-like" proteins. Cell. 1990 Nov 2;63(3):451–453. doi: 10.1016/0092-8674(90)90438-k. [DOI] [PubMed] [Google Scholar]
- Simpson R. T. Nucleosome positioning can affect the function of a cis-acting DNA element in vivo. Nature. 1990 Jan 25;343(6256):387–389. doi: 10.1038/343387a0. [DOI] [PubMed] [Google Scholar]
- Thomm M., Sherf B. A., Reeve J. N. RNA polymerase-binding and transcription initiation sites upstream of the methyl reductase operon of Methanococcus vannielii. J Bacteriol. 1988 Apr;170(4):1958–1961. doi: 10.1128/jb.170.4.1958-1961.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomm M., Wich G. An archaebacterial promoter element for stable RNA genes with homology to the TATA box of higher eukaryotes. Nucleic Acids Res. 1988 Jan 11;16(1):151–163. doi: 10.1093/nar/16.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weil C. F., Cram D. S., Sherf B. A., Reeve J. N. Structure and comparative analysis of the genes encoding component C of methyl coenzyme M reductase in the extremely thermophilic archaebacterium Methanothermus fervidus. J Bacteriol. 1988 Oct;170(10):4718–4726. doi: 10.1128/jb.170.10.4718-4726.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wich G., Hummel H., Jarsch M., Bär U., Böck A. Transcription signals for stable RNA genes in Methanococcus. Nucleic Acids Res. 1986 Mar 25;14(6):2459–2479. doi: 10.1093/nar/14.6.2459. [DOI] [PMC free article] [PubMed] [Google Scholar]